MitoDNA™ Red 680
MitoDNA™ Red 680 is a far-red mitochondrial DNA probe with deep tissue penetration that enables selective mtDNA imaging in live cells and tissues with minimal autofluorescence background.
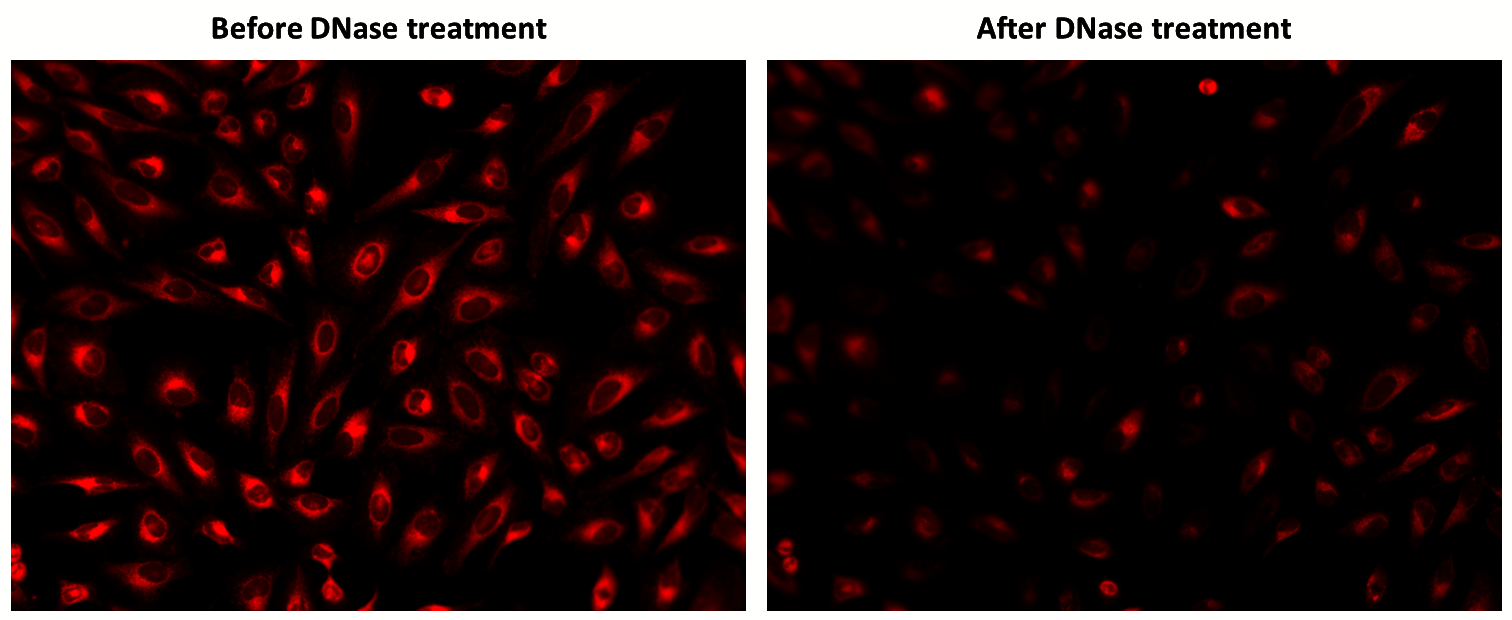
- mtDNA-Specific: Selectively binds to mtDNA without nuclear interference
- Live-Cell Compatibility: Easily permeates cells for real-time mtDNA tracking
- High Signal Clarity: Large Stokes shift ensures a strong signal-to-noise ratio for multiplex imaging
- Broad Research Applicability: Ideal for investigating mtDNA-linked diseases and mitochondrial function

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22688 | 1 mg | Price |
Physical properties
| Molecular weight | 486.44 |
| Solvent | DMSO |
Spectral properties
| Excitation (nm) | 597 |
| Emission (nm) | 681 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12171501 |
Instrument settings
| Fluorescence microscope | |
| Excitation | 600 nm |
| Emission | 680 nm |
| Recommended plate | Black wall/clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 15, 2026

