iFluor® 750 amine
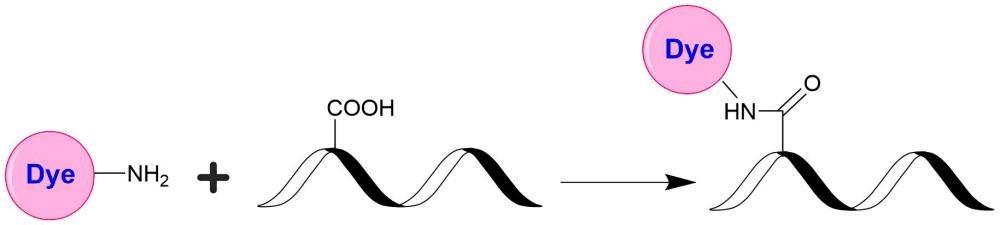
AAT Bioquest's iFluor® dyes are developed for labeling proteins, particularly antibodies. These dyes are optimized to have minimal fluorescence quenching effect on proteins and nucleic acids. Our iFluor® 750 dyes have fluorescence excitation and emission maxima of ~757 nm and ~779 nm, respectively, with good photostability. Our in-house comparable studies indicated that our iFluor® 750 dyes are significantly brighter than the corresponding Cy7® and Alexa Fluor® 750. These spectral characteristics make them a superior alternative to Cy7® and Alexa Fluor® 750 (Cy7® and Alexa Fluor® are the trademarks of GE Healthcare and Invitrogen). iFluor® 750 conjugates have been widely used in fluorescence animal imaging applications. The iFluor® 750 amine is stable and used for modifying carbonyl groups (e.g., aldehyde and carboxy groups).

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 1079 | 1 mg | Price |
Physical properties
| Molecular weight | 1047.07 |
| Solvent | DMSO |
Spectral properties
| Correction factor (260 nm) | 0.044 |
| Correction factor (280 nm) | 0.039 |
| Correction factor (565 nm) | 0.0250 |
| Correction factor (650 nm) | 0.1413 |
| Extinction coefficient (cm -1 M -1) | 275000 1 |
| Excitation (nm) | 757 |
| Emission (nm) | 779 |
| Quantum yield | 0.12 1 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12171501 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 14, 2026

