MitoDNA™ Blue 470
Product key features
- mtDNA-Specific: Selectively binds to mtDNA without nuclear interference
- Live-Cell Compatibility: Easily permeates cells for real-time mtDNA tracking
- High Signal Clarity: Large Stokes shift ensures a strong signal-to-noise ratio for multiplex imaging
- Broad Research Applicability: Ideal for investigating mtDNA-linked diseases and mitochondrial function
Product description
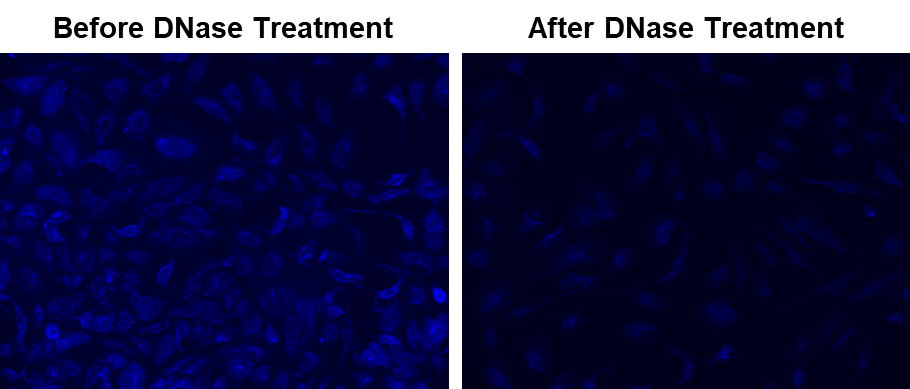
There are very few probes that can be effectively used to detect mitochondrial deoxyribonucleic acid (mtDNA). The common fluorescent DNA probes (such as DAPI, Hoechst or SYBR® Green) lack the specificity to target mitochondria. They predominantly stain nuclei. MitoDNA™ Blue 470 is a cell permeable dye that specifically stains mtDNA in live cells. It provides an efficient way of labeling mtDNA for the dynamic imaging of mtDNA in live cells. MitoDNA™ Blue 470 has a good Stokes Shift that gives great signal-to-noise ratio and can be easily used for multiplex staining in cells with other fluorescent imaging probes. mtDNA is a small circular DNA found within mitochondria present in the cytoplasm of a cell. This DNA is supplementary to the nucleic acid material found in the nucleus of each cell. The mtDNA codes for 37 genes that promote the proper functioning of some cells. The mitochondria synthesize adenosine triphosphate (ATP) through oxidative phosphorylation and encode information for the synthesis of enzymes, transfer ribonucleic acid (tRNA), and ribosomal RNA (rRNA). Disorders of mtDNA and mutations in its genes can predispose to health problems like age-related hearing loss, diabetes, and brain, heart, and liver failure, among other conditions. Moreover, mtDNA and its associated mitochondrial disorders can predispose people to different types of cancers including lymphomas, leukemias, and breast, intestine, liver and kidney tumors etc.
Example protocol
AT A GLANCE
Before using MitoDNA™ Blue 470 for the first time, allow it to thaw at room temperature. Then, briefly centrifuge it to collect the dried pellet.
Prepare cells in a growth medium.
Stain cells with MitoDNA™ Blue 470 working solution.
Incubate samples for 5 to 15 minutes at 37 °C.
Use a fluorescence microscope with a DAPI filter set to monitor fluorescence intensity.
PREPARATION OF STOCK SOLUTIONS
Unless otherwise noted, all unused stock solutions should be divided into single-use aliquots and stored at -20 °C after preparation. Avoid repeated freeze-thaw cycles
Prepare a 5 to 10 mM MitoDNA™ Blue 470 stock solution in DMSO. For example, add 150 μL of DMSO to the MitoDNA™ Blue 470 vial to create a 10 mM stock solution.
Note: Prepare a single aliquot of the unused MitoDNA™ Blue 470 stock solution and store it at ≤ -20 º C, protected from light. Avoid repeated freeze-thaw cycles.
PREPARATION OF WORKING SOLUTION
Prepare a 5 to 10 μM working solution by diluting the MitoDNA™ Blue 470 stock solution in Hanks' solution with 20 mM HEPES buffer (HHBS).
Note: For optimal results, use this solution within a few hours of preparation.
Note: Cover the working solution with foil or store it in a dark place to protect it from light.
SAMPLE EXPERIMENTAL PROTOCOL
Plate the cells in a 96-well plate with black walls and a clear bottom.
Remove the cell culture medium and add 100 µL of MitoDNA™ Blue 470 working solution directly to the cells.
Incubate the cells at 37°C for 5-15 minutes, protected from light.
Note: The concentration and incubation time of MitoDNA™ Blue 470 may vary depending on the cell line. Test different concentrations to determine the optimal dose.
Remove the dye working solution and wash the cells twice with HHBS buffer.
Add HHBS buffer and analyze the cells using a fluorescence microscope equipped with a DAPI filter set.
Spectrum
References
Authors: Raad, Georges and Tanios, Judy and Serdarogullari, Munevver and Bazzi, Marwa and Mourad, Youmna and Azoury, Joseph and Yarkiner, Zalihe and Liperis, Georgios and Fakih, Fadi and Fakih, Chadi
Journal: Journal of assisted reproduction and genetics (2024): 795-813
Authors: Llavanera, Marc and Mateo-Otero, Yentel and Viñolas-Vergés, Estel and Bonet, Sergi and Yeste, Marc
Journal: Journal of animal science and biotechnology (2024): 10
Authors: Geng, Qiang and Gao, Ruifang and Sun, Yuan and Chen, Shaofeng and Sun, Lili and Li, Wei and Li, Zhong and Zhao, Yu and Zhao, Feng and Zhang, Ying and Li, Anwen and Liu, Hongbin
Journal: Journal of assisted reproduction and genetics (2024)
Authors: Lozano, Manuel and McEachan, Rosemary R C and Wright, John and Yang, Tiffany C and Dow, Courtney and Kadawathagedara, Manik and Lepeule, Johanna and Bustamante, Mariona and Maitre, Lea and Vrijheid, Martine and Brantsæter, Anne Lise and Meltzer, Helle Margrete and Bempi, Vasiliki and Roumeliotaki, Theano and Thomsen, Cathrine and Nawrot, Tim and Broberg, Karin and Llop, Sabrina
Journal: The Science of the total environment (2024): 173014
Authors: Pereira-Martins, Diego A and Coelho-Silva, Juan L and Weinhäuser, Isabel and Franca-Neto, Pedro L and Silveira, Douglas R and Ortiz, César and Moreira-Aguiar, Amanda and Lima, Marinus M and Koury, Luisa C and de Melo, Raul A and Glória, Ana B and Fagundes, Evandro M and Lino, Bruno K and Pagnano, Katia and Bittencourt, Rosane and Nunes, Elenaide and Traina, Fabiola and Figueiredo-Pontes, Lorena and Keating, Armand and Tallman, Martin S and Ribeiro, Raul C and Dilon, Richard and Ganser, Arnold and Sanz, Miguel A and Berliner, Nancy and Valk, Peter and Löwenberg, Bob and Ottone, Tiziana and Noguera, Nelida I and Voso, Maria T and Paoloni, Francesca and Fazi, Paola and Ammatuna, Emanuele and Huls, Gerwin and Schuringa, Jan Jacob and Rego, Eduardo M and Lucena-Araujo, Antonio R
Journal: British journal of haematology (2023): 170-174