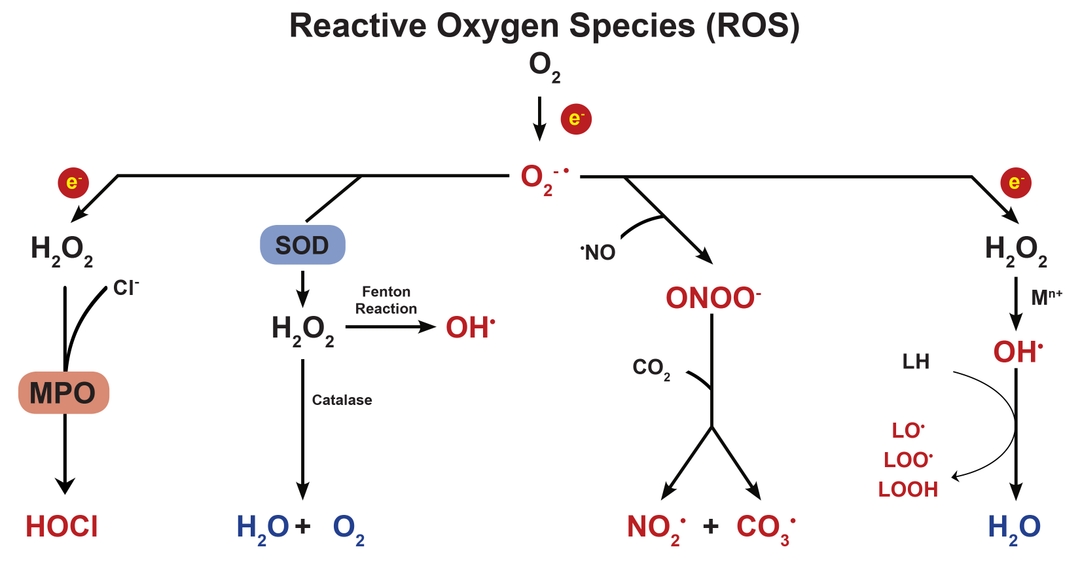
Selecting the right ROS probe
When generated in excess, ROS has long been thought to result in damage of cellular macromolecules such as DNA, lipids and proteins. Holistically, this is represented by a cell's oxidative stress state. Such states of stress have been linked to cellular processes such as apoptosis, and more macroscopically, play a role in the pathogenesis of many human diseases.
Because of its damaging effects, cells have several carefully regulated systems for managing excess ROS. The most well studied system is the glutathione-ascorbate cycle, which detoxifies H2O2 into H2O, using NADH and NADPH as electron donors. Other systems include enzymes such as superoxide dismutase, which catalyzes the dismutation of the superoxide anion (O2-) into O2 or H2O2, and catalase, which catalyzes the decomposition of H2O2 into H2O and O2.
While ROS has been extensively studied for their detrimental effect on cells, it is only more recently that studies have looked at the role of ROS in cell signaling. In controlled amounts, ROS has been shown to regulate gene activation. The specific mechanism by which this occurs is, however, still up for debate. It is possible that ROS binds to special receptors which initiate a signaling cascade leading up to gene regulation. It is also possible that ROS directly modifies the proteins in such a signaling cascade, perhaps by regulation of protein phosphorylation.
Due to its importance in biological systems, a plethora of tools have been developed to study ROS both in vitro and in vivo. The table below provides a summary of the most common tools as well as their targets.
|
Target |
Application |
Tools |
Catalog Number |
Principle |
|
Hydrogen peroxide (H2O2) a product of many enzymatic ROS scavenging pathways. The most well studied is superoxide dismutase activity, which catalyzes the reduction of superoxide anion to hydrogen peroxide. |
live cell |
DCFH-DA |
Probe enters cell wherein esterases cleave off diacetate group. Then DCFH is oxidized by hydrogen peroxide to DCF and emits green fluorescence upon excitation.[4] | |
|
Dihydrorhodamine 123 |
Probe passively permeates cell membrane. Oxidation by hydrogen peroxide yields rhodamine 123, which fluoresces blue upon excitation. | |||
|
OxiVision™ Blue |
Probe permeates cell and is oxidized by intracellular hydrogen peroxide. Generates fluorescence upon excitation. | |||
|
OxiVision™ Green | ||||
|
cell extract; solutions |
Amplite® Fluorimetric Hydrogen Peroxide Assay Kit (Red) |
Hydrogen peroxide dependent oxidation of ADHP (synonyms: 10-acetyl-3,7-dihydroxyphenoxazine, Amplex® Red) by horseradish peroxidase (HRP) converts ADHP to resorufin. Resorufin can be detected using colorimetric or fluorimetric methods. | ||
|
Amplite® Colorimetric Hydrogen Peroxide Assay Kit | ||||
|
Amplex™ Red Hydrogen Peroxide/Peroxidase Assay Kit |
Not available | |||
|
Amplite® Fluorimetric Hydrogen Peroxide Assay Kit (Infrared) |
Hydrogen peroxide dependent oxidation of Amplite IR by horseradish peroxidase (HRP) generates activated probe. Excitation emits a near-infrared fluorescent signal | |||
|
Superoxide anion (O2-) a by-product of aerobic metabolism, such as mitochondrial respiration (particularly Complex I and Complex III). |
live cell; cell extract; solutions |
Lucigenin |
Lucigenin is activated by conversion to lucigenin cation radical. Lucigenin cation radical reacts with superoxide anion to produce dioxetane intermediate, which decomposes to N-methylacridone. High energy electrons in N-methylacridone fall to lower energy state, resulting in luminescence.[5] | |
|
Coelenterazine |
Oxidation by superoxide anion results in an excited electron state. Upon decay to ground state electron configuration, photons are released as luminescence.[8] | |||
|
Luminol |
Superoxide-dependent enzyme catalyzed oxidation of luminol results in luminescence.[6] | |||
|
Hydroethidine |
Probe passively permeates intact cells and localizes in the mitochondria. Probe is activated through oxidation by superoxide. The activated probe intercalates with DNA and, upon excitation, fluoresces. DNA binding is necessary for strong fluorescence signal. For hydroethidine, activated probe is ethidium (ie. same active species as DNA stain ethidium bromide).[3,7,12] | |||
|
MitoSox™ Red |
Not available | |||
|
MitoROS™ 580 | ||||
|
MitoROS™ 520 |
Probe readily passes through intact cell membranes whereupon it localizes in mitochondria. It is then oxidized by superoxide. Upon excitation, it releases a green fluorescence. | |||
|
Hydroxyl radical (•OH) can be generated when superoxide anions react with transition metals. Extremely reactive. Can react with hydrogen on DNA backbone, resulting in strand breakage. |
live cell; cell extract; solutions |
MitoROS™ OH580 |
Probe is able to freely enter live cells wherein it becomes oxidized specifically by free hydroxyl radicals. Upon excitation, oxidized probe fluoresces red. | |
|
Total ROS includes hydrogen peroxide, superoxide anion, hydroxyl radical, singlet oxygen, nitric oxide, butyl peroxide and hypochlorous acid |
live cell; cell extract; solutions |
ROS™ Brite 570 |
Probe passively permeates intact cell membranes. Once inside the cell, probe is oxidized by intracellular ROS. Probe can also be oxidized by ROS in solution for cell extract assays. Upon excitation, probe emits a fluorescent signal. | |
|
ROS™ Brite 670 | ||||
|
ROS™ Brite 700 | ||||
|
Amplite® ROS Green |
|
Target |
Tool |
Catalog Number |
Principle |
|
SOD dismutase (SOD) |
Amplite® Colorimetric Superoxide Dismutase (SOD) Assay Kit |
First uses xanthine oxidase (XO) to convert xanthine into hydrogen peroxide and uric acid while simultaneously catalyzing the reduction of molecular oxygen (O2) into superoxide anion (O2-). Then uses competitive inhibition of superoxide dismutation by ReadiView™ SOD560 to quantify superoxide dismutase activity. Decrease in absorbance of ReadiView™ SOD560 is directly proportional to SOD activity. | |
|
Catalase |
Amplite® Fluorimetric Catalase Assay Kit (Red) |
Competitive inhibition assay. Amplite® Red probe is activated through oxidation by hydrogen peroxide. Thus, probe competes with catalase for hydrogen peroxidase substrate. Amplite Red absorbance is inversely proportional to catalase activity. | |
|
Glutathione (GSH) |
Thiolite™ Green |
Probe becomes activated after reaction with glutathione. Upon excitation, probe releases a green fluorescence. | |
|
Amplite® Fluorimetric Glutathione GSH/GSSG Ratio Assay Kit |
Uses Thiolite™ Green probe, which becomes activated after reaction with glutathione (GSH), to quantify glutathione. Oxidized glutathione (GSSG) is determined by measuring GSH concentration before and after enzyme-catalyzed reduction of GSSG to GSH. GSSG concentration is calculated by subtracting initial GSH (before enzyme reaction) from total GSH (after enzyme reaction). | ||
|
Glutathione peroxidase (GPx) |
Amplite® Fluorimetric Glutathione Peroxidase Assay Kit (Blue) |
Enzymatic cycling assay. Glutathione peroxidase (GPx) catalyzes the oxidation of glutathione from GSH to GSSG. Glutathione reductase (GR) then catalyzes the reduction of GSSG back into GSH with the coenzyme NADPH, which is oxidized to NADP+. The Quest Fluor™ NADP Probe then quantifies the level of NADP+ which is directly proportional to the original GPx activity. | |
|
Ascorbate |
Amplite® Fluorimetric Ascorbic Acid Assay Kit (Blue) |
Relies on the dehydrogenation of ascorbic acid to dehydroascorbic acid (DHA) by an enzyme-catalyzed reaction. Resulting DHA is quantified by Ascorbrite Blue probe. | |
|
NAD+/NADH |
Amplite® NAD/NADH Kits (assorted) |
15273, 15258, 15275, 15280, 15263, 15257, 15261, 15262, 15291, 15290, 15259, 15271 |
Typically, uses a NAD+ specific probe to quantify NAD+ concentration. NAD+/NADH ratio is determined by enzymatic cycling assay. |
|
NADPH |
Amplite® NADPH Kits (assorted) |
15274, 15272, 15260, 15276, 15264, 15262, 15259, 15291, 15290 |
Typically, uses a NADPH specific probe to quantify NADPH concentration. NADP/NADPH ratio is determined by enzymatic cycling assay. |
References
- Apel, Klaus, and Heribert Hirt. "Reactive oxygen species: metabolism, oxidative stress, and signal transduction." Rev. Plant Biol.55 (2004): 373-399.
- Bindokas, Vytautas P., et al. "Superoxide production in rat hippocampal neurons: selective imaging with hydroethidine."Journal of Neuroscience4 (1996): 1324-1336.
- Kalyanaraman, Balaraman, et al. "Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations."Free Radical Biology and Medicine1 (2012): 1-6.
- LeBel, Carl P., Harry Ischiropoulos, and Stephen C. Bondy. "Evaluation of the probe 2', 7'-dichlorofluorescin as an indicator of reactive oxygen species formation and oxidative stress."Chemical research in toxicology2 (1992): 227-231.
- Li, Yunbo, et al. "Validation of lucigenin (bis-N-methylacridinium) as a chemilumigenic probe for detecting superoxide anion radical production by enzymatic and cellular systems."Journal of Biological Chemistry4 (1998): 2015-2023.
- Misra, Hara P., and Pamela M. Squatrito. "The role of superoxide anion in peroxidase-catalyzed chemiluminescence of luminol."Archives of biochemistry and biophysics1 (1982): 59-65.
- Mukhopadhyay, Partha, et al. "Simple quantitative detection of mitochondrial superoxide production in live cells."Biochemical and biophysical research communications1 (2007): 203-208.
- Münzel, Thomas, et al. "Detection of superoxide in vascular tissue."Arteriosclerosis, thrombosis, and vascular biology11 (2002): 1761-1768.
- Murphy, Michael P. "How mitochondria produce reactive oxygen species."Biochemical Journal1 (2009): 1-13.
- Thannickal, Victor J., and Barry L. Fanburg. "Reactive oxygen species in cell signaling."American Journal of Physiology-Lung Cellular and Molecular Physiology6 (2000): L1005-L1028.
- Turrens, Julio F. "Mitochondrial formation of reactive oxygen species."The Journal of physiology2 (2003): 335-344.
- Zielonka, Jacek, and B. Kalyanaraman. "Hydroethidine-and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth."Free Radical Biology and Medicine48.8 (2010): 983-1001.