ReadiLink™ Rapid trFluor™ Eu Antibody Labeling Kit *Microscale Optimized for Labeling 50 ug Antibody Per Reaction*
Many biological compounds present in cells, serum or other biological fluids are naturally fluorescent, and thus the use of conventional, prompt fluorophores leads to serious limitations in assay sensitivity due to the high background caused by the autofluorescence of the biological molecules to be assayed. The use of long-lived fluorophores combined with time-resolved detection (a delay between excitation and emission detection) minimizes prompt fluorescence interferences. Our TR Fluor™ Eu probes enable time-resolved fluorometry (TRF) for the assays that require high sensitivity. These TR Fluor™ Eu probes have large Stokes shifts and extremely long emission half-lives when compared to more traditional fluorophores such as Alexa Fluor or cyanine dyes. Compared to the other TRF compounds, our TR Fluor™ Eu probes have relatively high stability, high emission yield and ability to be linked to biomolecules. Moreover, our TR Fluor™ Eu probes are insensitive to fluorescence quenching when conjugated to biological polymers such as antibodies. 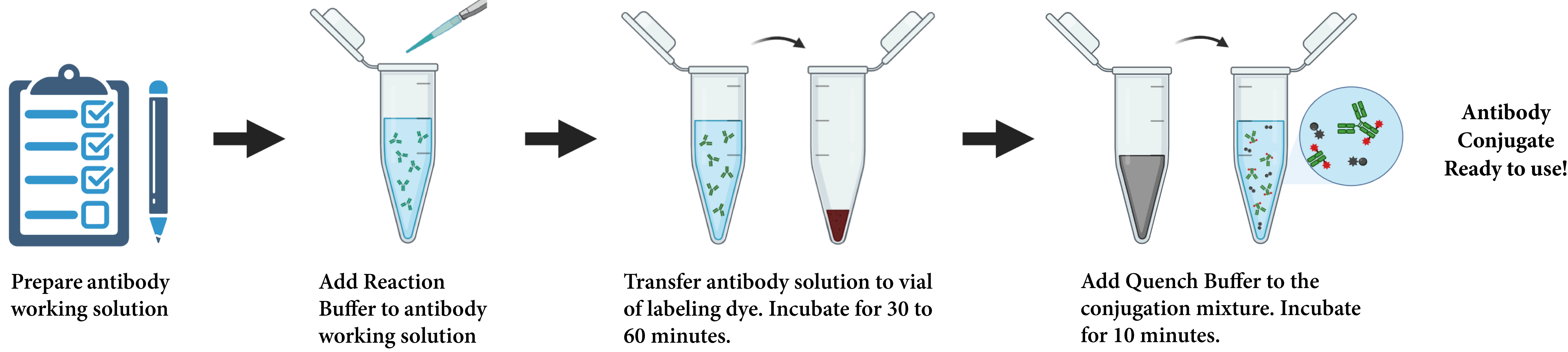

Figure 1. Overview of the ReadiLink™ Rapid Antibody Labeling protocol. In just two simple steps, and with no purification necessary, covalently label microgram amounts of antibodies in under an hour.
Example protocol
AT A GLANCE
Important
Upon receipt, store labelling dye (Component A) at -20 °C, kept from light and moisture. Store other components at room temperature. When stored properly, the kit components should be stable for six months.Note Do not freeze Reaction Buffer (Component B) and Spin Column (Component C).
Note Warm all the components and centrifuge the vials briefly before opening, and immediately prepare the required solutions before starting your conjugation. The following SOP is an example for labeling anti-HDAC IgG antibody.
PREPARATION OF WORKING SOLUTION
Protein Working Solution (Solution A)
For labeling 50 µg of protein (assuming the target protein concentration is 1 mg/mL), mix 1.5 µL (3% of the total reaction volume) of Reaction Buffer (Component B) with 50 µL of the target protein solution.Note If you have a different protein concentration, adjust the protein volume accordingly to make ~50 µg of protein available for your labeling reaction.
Note For labeling 100 µg of protein (assuming the target protein concentration is 1 mg/mL), mix 3 µL (3% of the total reaction volume) of Reaction Buffer (Component B) with 100 µL of the target protein solution.
Note The protein should be dissolved in 1X phosphate buffered saline (PBS), pH 7.2 - 7.4; if the protein is dissolved in glycine buffer, it must be dialyzed against 1X PBS, pH 7.2 - 7.4, or use Amicon Ultra-0.5, Ultracel-10 Membrane, 10 kDa (cat# UFC501008 from Millipore) to remove free amines or ammonium salts (such as ammonium sulfate and ammonium acetate) that are widely used for protein precipitation.
Note Impure antibodies or antibodies stabilized with bovine serum albumin (BSA) or gelatin will not be labeled well.
Note The conjugation efficiency is significantly reduced if the protein concentration is less than 1 mg/mL. For optimal labeling efficiency the final protein concentration range of 1-2 mg/mL is recommended.
SAMPLE EXPERIMENTAL PROTOCOL
Run conjugation reaction
- Add the protein solution (Solution A) to ONE vial of labeling dye (Component A), and mix them well by repeatedly pipetting for a few times or vortex the vial for a few seconds.
Note If labeling 100 µg of protein, use both vials (Component A) of labeling dye by dividing the 100 of µg protein into 2 x 50 µg of protein and reacting each 50 µg of protein with one vial of labeling dye. Then combine both vials for the next step. - Keep the conjugation reaction mixture at room temperature for 30 - 60 minutes.
Note The conjugation reaction mixture can be rotated or shaken for a longer time if desired.
Prepare spin column for sample purification
- Invert the Spin Column (Component C) several times to resuspend the settled gel and remove any bubbles.
- Snap off the tip and place the column in the Washing Tube (2 mL, Component D). Remove the cap to allow the excess packing buffer to drain by gravity to the top of the gel bed. If column does not begin to flow, push cap back into column and remove it again to start the flow. Discard the drained buffer, and then place the column back into the Washing Tube. However, centrifuge immediately if the column is placed into a 12 x 75 mm test tube (not provided).
- Centrifuge for 1 min in a swinging bucket centrifuge at 1,000 x g (see Centrifugation Notes) to remove the packing buffer. Discard the buffer.
- Apply 1 - 2 mL 1X PBS (pH 7.2 - 7.4) to the column. After each application of PBS, let the buffer drain out by gravity, or centrifuge the column for 1 min to remove the buffer. Discard the buffer from the collection tube. Repeat this process for 3 - 4 times.
- Centrifuge for 2 minutes in a swinging bucket centrifuge at 1,000 x g (see Centrifugation Notes) to remove the packing buffer. Discard the buffer.
Purify the conjugation
- Place the column in a clean Collecting Tube (1.5 mL, Component E). Carefully load the sample (50–100 µL) directly to the center of the column.
- After loading the sample, add 1X PBS (pH 7.2-7.4) to make the total volume of 110 µL. Centrifuge the column for 5 min at 1,000 x g, and collect the solution that contains the desired dye-labeled protein.
Note For immediate use, the dye-protein conjugate needs to be diluted with the staining buffer before being aliquoted for multiple uses. For longer term storage, the dye-protein conjugate solution needs to be concentrated or freeze dried.
Storage of Protein Conjugate
The protein conjugate should be stored at >0.5 mg/mL in the presence of a carrier protein (e.g., 0.1% bovine serum albumin). The conjugate solution can be stored at 4°C for two months without significant change when stored in the presence of 2 mM sodium azide and kept from light. For longer storage, the protein conjugates can be lyophilized or divided into single-used aliquots and stored at ≤–20°C.Centrifugation Notes
Spin Column (Component C) can fit into 2 mL microcentrifuge tubes or 12 x 75 mm test tubes for sample collection during centrifugation. Use the 2 mL microtubes provided with the columns for the initial column equilibration step. Swinging bucket centrifuges capable of generating a minimum force of 1,000 x g are suitable for Bio-Spin column use. The gravitational force created at a particular revolution speed is a function of the radius of the microcentrifuge rotor. Consult the swinging bucket centrifuge instruction manual for the information about conversion from revolutions per minute (RPM) to centrifugal or g-force. Alternatively, use the following equation to calculate the speed in RPM required to reach the gravitational force of 1,000 x g. RCF (x g) = (1.12 x 10-5)×(RPM)×2×r (RCF is the relative centrifugal force, r is the radius in centimeters measured from the center of the rotor to the middle of the Bio-Spin column, and RPM is the speed of the rotor).Spectrum
Product family
| Name | Excitation (nm) | Emission (nm) | Correction Factor (260 nm) | Correction Factor (280 nm) |
| ReadiLink™ Rapid trFluor™ Tb Antibody Labeling Kit *Microscale Optimized for Labeling 50 ug Antibody Per Reaction* | 333 | 544 | 0.942 | 0.797 |
References
View all 61 references: Citation Explorer
Development of a time-resolved fluorescence resonance energy transfer assay for cyclin-dependent kinase 4 and identification of its ATP-noncompetitive inhibitors
Authors: Lo MC, Ngo R, Dai K, Li C, Liang L, Lee J, Emkey R, Eksterowicz J, Ventura M, Young SW, Xiao SH.
Journal: Anal Biochem (2012): 368
Authors: Lo MC, Ngo R, Dai K, Li C, Liang L, Lee J, Emkey R, Eksterowicz J, Ventura M, Young SW, Xiao SH.
Journal: Anal Biochem (2012): 368
Time-Resolved Fluorescence Resonance Energy Transfer as a Versatile Tool in the Development of Homogeneous Cellular Kinase Assays
Authors: Saville L, Spais C, Mason JL, Albom MS, Murthy S, Meyer SL, Ator MA, Angeles TS, Husten J.
Journal: Assay Drug Dev Technol. (2012)
Authors: Saville L, Spais C, Mason JL, Albom MS, Murthy S, Meyer SL, Ator MA, Angeles TS, Husten J.
Journal: Assay Drug Dev Technol. (2012)
Oligomerization of the serotonin(1A) receptor in live cells: a time-resolved fluorescence anisotropy approach
Authors: Paila YD, Kombrabail M, Krishnamoorthy G, Chattopadhyay A.
Journal: J Phys Chem B (2011): 11439
Authors: Paila YD, Kombrabail M, Krishnamoorthy G, Chattopadhyay A.
Journal: J Phys Chem B (2011): 11439
A homogeneous single-label time-resolved fluorescence cAMP assay
Authors: Martikkala E, Rozw and owicz-Jansen A, Hanninen P, Petaja-Repo U, Harma H.
Journal: J Biomol Screen (2011): 356
Authors: Martikkala E, Rozw and owicz-Jansen A, Hanninen P, Petaja-Repo U, Harma H.
Journal: J Biomol Screen (2011): 356
Time-resolved fluorescence resonance energy transfer (TR-FRET) to analyze the disruption of EGFR/HER2 dimers: a new method to evaluate the efficiency of targeted therapy using monoclonal antibodies
Authors: Gaborit N, Larbouret C, Vallaghe J, Peyrusson F, Bascoul-Mollevi C, Crapez E, Azria D, Chardes T, Poul MA, Mathis G, Bazin H, Pelegrin A.
Journal: J Biol Chem (2011): 11337
Authors: Gaborit N, Larbouret C, Vallaghe J, Peyrusson F, Bascoul-Mollevi C, Crapez E, Azria D, Chardes T, Poul MA, Mathis G, Bazin H, Pelegrin A.
Journal: J Biol Chem (2011): 11337
Page updated on April 15, 2025



