Cell Meter™ Fluorescence Gap Junction Tracing Kit
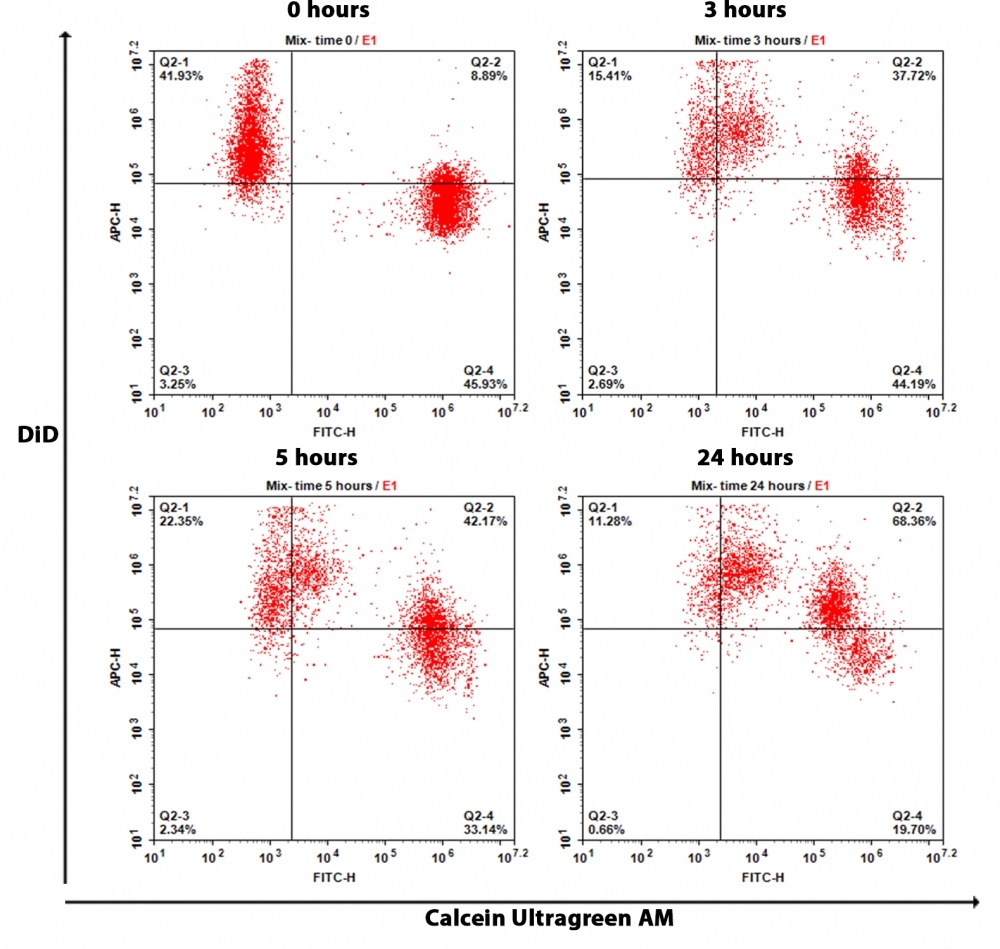
Gap junctions are specialized intercellular membrane channels constituted with connexin that selectively facilitate the passage of small molecules of <1.5 kD across cells. They are tightly regulated by voltage, growth factors, cAMP, and retinoids, and they are modulated by phosphorylation. Cell Meter™ Fluorescence Gap Junction Tracing Kit provides a reliable and robust assay for the in vitro determination of gap junction function. The method is noninvasive. The cell population under study is divided such that one fraction is loaded with a lipophilic cell plasma membrane permeable dye, Calcein UltraGreen™ AM, that is hydrolyzed upon cellular uptake by cytoplasmic esterases to yield Calcein UltraGreen, a highly fluorescent and well-retained and membrane-impermeable molecule. The other fraction is loaded DiD, which is a lipophilic membrane dye that diffuses laterally to stain the entire cell membrane in deep red fluorescence upon incorporation into membranes. The two fractions are mixed and incubated under coculture conditions. Calcein UltraGreen is transferred to the DiD-stained cells through gap junctions. The assessment of this uptake can be monitored by fluorescence imaging or flow cytometry.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 23600 | 100 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12171501 |
Instrument settings
| Flow cytometer | |
| Excitation | 488 nm and 640 nm laser |
| Emission | 530/30 nm and 660/20 nm filters |
| Instrument specification(s) | FITC and APC channel |
| Fluorescence microscope | |
| Excitation | FITC and Cy5 filter sets |
| Emission | FITC and Cy5 filter sets |
| Recommended plate | Black wall/clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 23, 2026
