Principles of Common Affinity Purification Formats
by K Chico, Jessica Piczon
.png&w=828&q=100)
Affinity purification, also commonly called affinity chromatography, is a group of diverse and powerful methods used to separate and purify molecule(s) from complex biological samples. It is a fast, often single step, process that enables purification in order of several hundred-fold or higher.
Affinity purification is based on the highly specific biological interactions between at least two molecules, which may be an enzyme and substrate, a receptor and ligand, or even an antibody and antigen. Generally, it is important to have some level of knowledge about the interaction of the two molecules as the reaction is, ideally, reversible. Knowledge of the binding interaction also helps determine how to select an appropriate affinity ligand and protocol. In simple terms, affinity purification separates target proteins based on their biological function, beneficial for concentrating molecules that may be present at extremely low levels within a sample.
First the sample in solution (mobile phase) is incubated with the affinity support (stationary phase). The mobile phase is normally composed of the cell lysate, serum, and/or growth medium, while the affinity support is some sort of solid support. This, too, depends, as there are many commercial affinity matrices available that often come pre-coupled to a ligand. Such ligands become immobilized to the solid support material, generally, by one of two means: through the formation of covalent bonds between functional groups and the matrix, or through indirect coupling.
Next, the sample undergoes incubation in conditions that favor binding of the target molecule in the sample with the immobilized ligand. After the target molecules are captured, the matrix is washed. In washing, compounds in the sample that have little to zero interaction with the ligand become separated from the stationary phase. Additionally, all target molecules remain stuck to the matrix due to their high affinity to the ligands present. After non-target molecules are washed away, the target molecules are released through an elution step. The affinity purification reaction results in the recovery of a highly purified, extremely concentrated target protein that can be collected for direct analysis and/or downstream applications.
The success of an affinity purification depends largely on the selection of the ligand, or binding agent, that is immobilized in the solid support. Biological ligands (e.g. antibodies, serum proteins, and lectins) can be used as well as non-biological ligands which are synthetic agents or inorganic molecules (e.g., boronic acid, metal chelates, and triazine dyes). High-specificity ligands are those that retain very few closely related targets and are common when the goal of the reaction is isolation and/or separation. Some examples of high-specificity ligands are antibodies for binding antigens, substrates, or inhibitors for separating enzymes, and single-strand nucleic acids for targeting specific DNA or RNA sequences. Alternatively, general ligands are used to isolate a class of related molecules or structurally similar targets. This subclass contains lectins and boronates for binding carbohydrate-containing agents, dyes for isolating specific residues on enzymes and proteins, and protein A or G for binding immunoglobulins.
The choice of the solid support matrix used should also be another major consideration. This solid support matrix should have low nonspecific binding for the sample and must be easy to chemically modify for ligand attachment. The solid support should also be able to withstand the necessary pressures and flow rates that will be used in the affinity purification protocol. Agarose and cellulose, both composed of organic polysaccharide chains, are often the major component used in traditional affinity columns. In comparison, silica particles, modified polystyrene supports, silica and other organic monoliths are commonly used in high performance techniques. More recently, functionalized magnetic nanoparticles as an alternative to the column format.
Regardless, all techniques rely on the nonspecific immobilization of the affinity ligand to the solid support. Noncovalent immobilization refers to the biospecific adsorption of the binding between two affinity ligands: the first is bound to the solid support and the second uses the first affinity ligand as a target. In covalent immobilization, ligands are chemically attached to a solid support that has been previously activated or functionalized. Covalent immobilization uses chemical functional groups to facilitate this reaction (e.g., amine, carboxyl, sulfhydryl, hydroxyl, or aldehyde groups). Affinity ligands can also be immobilized through other means, like entrapment or encapsulation, into the solid support.
Additionally, the buffers used for the affinity purification reactions should have an appropriate pH and ionic strength to promote binding between the immobilized ligand and target. The elution buffer must be strong enough to disrupt the binding of the target protein with the ligand-support. Elution is commonly performed through changing the pH, ionic strength, or adding a small amount of organic solvent in the mobile phase. Sometimes a competing agent may be added into the mobile phase instead to displace the target from the column by inhibiting the binding reaction. Though this method is often slow, it is quite useful for gently removing the active target from the support matrix.
The most common formation for affinity purification is performed through traditional column chromatography. This scheme uses affinity columns as the matrices which immobilize the ligands. Buffers used typically have pHs and ionic strengths that mimic the native environments of the ligand and its protein target. Similarly, high performance affinity column (HPAC) combines this traditional method with high performance liquid chromatography. HPAC supports are typically crafted using narrow-diameter silica or glass particles, perfusion-based materials, and monolithic supports. HPAC offers faster test times, better precision, and automation, making HPAC highly suitable for use in analytical applications. Affinity chromatography approaches can also be combined with detection methods through absorbance, fluorescence, or mass spectrometry.
Another affinity purification format common to biomedical analysis is immunoaffinity chromatography. In immunoaffinity chromatography, a specific analyte or group thereof is separated from the sample by a column that contains an immobilized affinity ligand. The immobilized ligand is often an antibody, and to a lesser extent a boronate or lectin. This technique has commonly been used in drug analysis to measure the fraction of drug components compared with hormones or protein levels in serum. In comparison, affinity depletion aims to remove abundant compounds from a sample to analyze the compounds that are eluted, and not the ones that bind to the column.
Affinity depletion is used widely in proteomics, and usually aims to remove highly abundant proteins (e.g., HSA and/or IgG) from serum prior to the analysis of the lower abundance proteins in the sample. Other methods of affinity purification can be divided based on the type of reversible affinity-ligand reaction including bioaffinity, immobilized metal-ion affinity, and dye-ligand and biomimetic affinity chromatography.
Chapter 26 Affinity Chromatography: General Methods
Chapter 26 PHARMACEUTICAL AND BIOMEDICAL APPLICATIONS OF AFFINITY CHROMATOGRAPHY: RECENT TRENDS AND DEVELOPMENTS
Affinity Chromatography: A Review of Trends and Developments over the Past 50 Years
Affinity chromatography: general methods
Chapter 12 - Affinity chromatography
Original created on February 3, 2024, last updated on February 3, 2024
Tagged under: Affinity, Purification
.png&w=828&q=100)
Simplified affinity chromatography column interactions, from initial sample addition and incubation, through washing steps and eventual elution of purified target protein. Figure made in Biorender.
Affinity purification is based on the highly specific biological interactions between at least two molecules, which may be an enzyme and substrate, a receptor and ligand, or even an antibody and antigen. Generally, it is important to have some level of knowledge about the interaction of the two molecules as the reaction is, ideally, reversible. Knowledge of the binding interaction also helps determine how to select an appropriate affinity ligand and protocol. In simple terms, affinity purification separates target proteins based on their biological function, beneficial for concentrating molecules that may be present at extremely low levels within a sample.
How Does Affinity Purification Work?
First the sample in solution (mobile phase) is incubated with the affinity support (stationary phase). The mobile phase is normally composed of the cell lysate, serum, and/or growth medium, while the affinity support is some sort of solid support. This, too, depends, as there are many commercial affinity matrices available that often come pre-coupled to a ligand. Such ligands become immobilized to the solid support material, generally, by one of two means: through the formation of covalent bonds between functional groups and the matrix, or through indirect coupling.
Next, the sample undergoes incubation in conditions that favor binding of the target molecule in the sample with the immobilized ligand. After the target molecules are captured, the matrix is washed. In washing, compounds in the sample that have little to zero interaction with the ligand become separated from the stationary phase. Additionally, all target molecules remain stuck to the matrix due to their high affinity to the ligands present. After non-target molecules are washed away, the target molecules are released through an elution step. The affinity purification reaction results in the recovery of a highly purified, extremely concentrated target protein that can be collected for direct analysis and/or downstream applications.
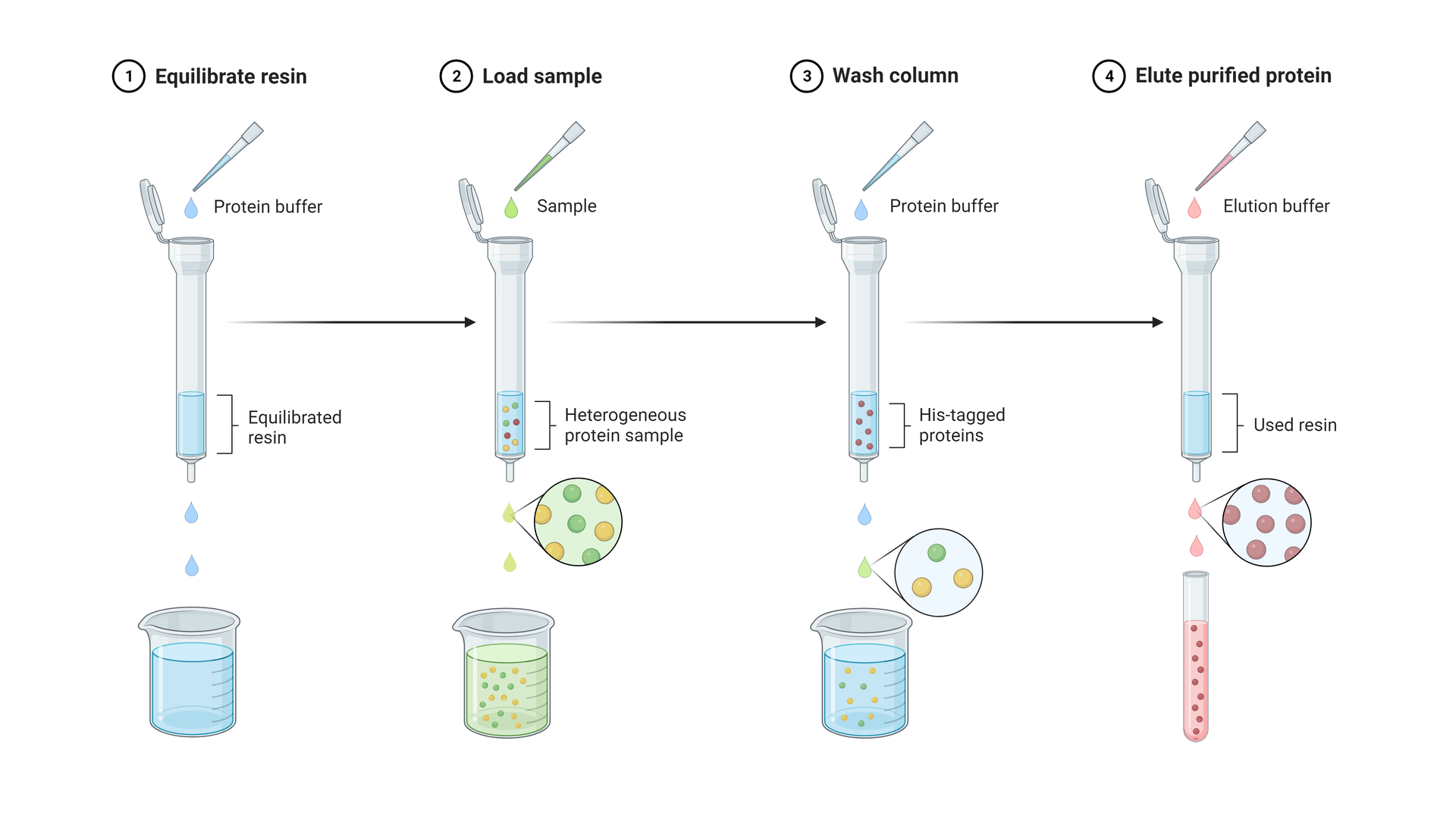
Components of Affinity Purification

Simplified steps of a basic affinity purification procedure, in this case using a His-tagged antibody for the affinity interaction. Figure made in Biorender.
Table 1. Protein A for Affinity Purification
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 11041 | MegaWox™ polyHRP-Protein A Conjugate | 1 mg |
| 55000 | Protein A-Agarose Resin | 1 mL |
| 55005 | Protein A-Agarose Resin | 5 mL |
The choice of the solid support matrix used should also be another major consideration. This solid support matrix should have low nonspecific binding for the sample and must be easy to chemically modify for ligand attachment. The solid support should also be able to withstand the necessary pressures and flow rates that will be used in the affinity purification protocol. Agarose and cellulose, both composed of organic polysaccharide chains, are often the major component used in traditional affinity columns. In comparison, silica particles, modified polystyrene supports, silica and other organic monoliths are commonly used in high performance techniques. More recently, functionalized magnetic nanoparticles as an alternative to the column format.
Regardless, all techniques rely on the nonspecific immobilization of the affinity ligand to the solid support. Noncovalent immobilization refers to the biospecific adsorption of the binding between two affinity ligands: the first is bound to the solid support and the second uses the first affinity ligand as a target. In covalent immobilization, ligands are chemically attached to a solid support that has been previously activated or functionalized. Covalent immobilization uses chemical functional groups to facilitate this reaction (e.g., amine, carboxyl, sulfhydryl, hydroxyl, or aldehyde groups). Affinity ligands can also be immobilized through other means, like entrapment or encapsulation, into the solid support.
Additionally, the buffers used for the affinity purification reactions should have an appropriate pH and ionic strength to promote binding between the immobilized ligand and target. The elution buffer must be strong enough to disrupt the binding of the target protein with the ligand-support. Elution is commonly performed through changing the pH, ionic strength, or adding a small amount of organic solvent in the mobile phase. Sometimes a competing agent may be added into the mobile phase instead to displace the target from the column by inhibiting the binding reaction. Though this method is often slow, it is quite useful for gently removing the active target from the support matrix.
General Formats for Affinity Purification

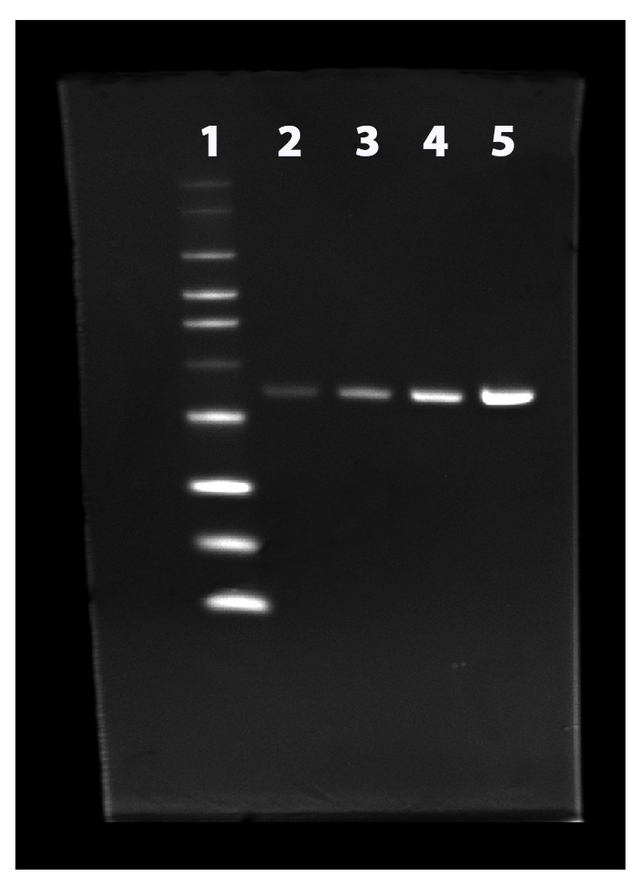
Two-fold dilution series of His-tagged annexin V were separated on a NuPAGE® 4-12% Bis-Tris gel and stained with the ProLite™ His-Tag Protein Gel Staining Kit according to standard protocols. Lane 1: His-tagged protein ladder, Lane 2 to 5: two-fold dilution of His-tagged annexin V.
Another affinity purification format common to biomedical analysis is immunoaffinity chromatography. In immunoaffinity chromatography, a specific analyte or group thereof is separated from the sample by a column that contains an immobilized affinity ligand. The immobilized ligand is often an antibody, and to a lesser extent a boronate or lectin. This technique has commonly been used in drug analysis to measure the fraction of drug components compared with hormones or protein levels in serum. In comparison, affinity depletion aims to remove abundant compounds from a sample to analyze the compounds that are eluted, and not the ones that bind to the column.
Affinity depletion is used widely in proteomics, and usually aims to remove highly abundant proteins (e.g., HSA and/or IgG) from serum prior to the analysis of the lower abundance proteins in the sample. Other methods of affinity purification can be divided based on the type of reversible affinity-ligand reaction including bioaffinity, immobilized metal-ion affinity, and dye-ligand and biomimetic affinity chromatography.
Products
Table 2. IDA ligands for Immobilized metal affinity chromatography (IMAC)
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 12630 | Biotin-X IDA | 1 mg |
| 12631 | IDA maleimide | 5 mg |
| 12632 | IDA succinimidyl ester | 5 mg |
Table 3. HIS Lite™ products
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 12610 | HIS Lite™ Cy3 Bis NTA-Ni Complex | 1 mg |
| 12613 | HIS Lite™ Cy5 Bis NTA-Ni Complex | 1 mg |
| 12615 | HIS Lite™ OG488-Tris NTA-Ni Complex | 100 µg |
| 12617 | HIS Lite™ iFluor® 568 Tris NTA-Ni Complex | 100 µg |
| 12618 | HIS Lite™ iFluor™ 647 Tris NTA-Ni Complex | 100 µg |
| 12619 | HIS Lite™ Cy5 Tris NTA-Ni Complex | 100 µg |
| 12620 | HIS Lite™ Cy3 Tris NTA-Ni Complex | 100 µg |
References
Chapter 26 Affinity Chromatography: General Methods
Chapter 26 PHARMACEUTICAL AND BIOMEDICAL APPLICATIONS OF AFFINITY CHROMATOGRAPHY: RECENT TRENDS AND DEVELOPMENTS
Affinity Chromatography: A Review of Trends and Developments over the Past 50 Years
Affinity chromatography: general methods
Chapter 12 - Affinity chromatography
Original created on February 3, 2024, last updated on February 3, 2024
Tagged under: Affinity, Purification