Streptavidin
Streptavidin is a biotin-binding protein with exceptional affinity, widely used as a versatile tool for molecular detection, purification, and signal amplification in research and diagnostics.
- Ultra-High Biotin Affinity: Kd = 10⁻¹⁵ M, among strongest non-covalent interactions
- Tetrameric Structure: Four biotin binding sites per 60 kDa protein
- pH/Temperature Stable: Active from pH 3–10 and up to 70°C
- Low Nonspecific Binding: pI ~5–6 reduces background in immunoassays

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 16885 | 1 mg | Price | |
| 16886 | 10 mg | Price |
Overview
Streptavidin, purified from the Streptomyces avidinii bacterium, is a cornerstone reagent in molecular biology, biochemistry, and biotechnology, employed for its extraordinarily high affinity and specificity toward biotin (vitamin B7). This noncovalent interaction, often cited as one of the strongest known in nature, underpins a multitude of applications in diagnostics, protein and nucleic acid purification, biosensing, and cutting-edge analytical methods. Streptavidin's unique properties—ranging from its structural robustness to its remarkable ligand-binding kinetics—have made it a go-to solution for both routine assays and highly specialized research protocols. Over the past several decades, extensive studies and refinements have broadened its utility, enabling sophisticated workflows in protein engineering, single-molecule detection, high-throughput screening, and beyond. Such enduring versatility reflects not only streptavidin's intrinsic strengths but also the depth of scientific understanding developed through continuous study and innovation.
Molecular Properties and Structural Insights
The streptavidin molecular weight is approximately 52.8 kDa in its native tetrameric form, each subunit spanning about 13 kDa (Hermanson, 2008; Chaiet and Wolf, 1964). Its amino acid composition (roughly 159 residues per subunit) yields a polypeptide devoid of glycosylation, ensuring a neutral overall charge at physiological pH. The absence of carbohydrate moieties reduces nonspecific binding and enhances reproducibility, a critical factor when optimizing complex assays.
X-ray crystallography and streptavidin PDB structures (e.g., PDB ID: 1STP) reveal a β-barrel fold in each monomer, creating a high-affinity binding site that perfectly accommodates biotin's ureido ring and valeric acid side chain. The streptavidin structure, stabilized by hydrogen bonds and hydrophobic interactions, assembles into a stable tetramer that binds four biotin molecules. Each of these four binding sites show almost negligible structural rearrangement upon biotin binding, reflecting a near-perfect fit. (Gonzalez et al., 2000; Laitinen et al., 2006)
Notably, the dissociation constant (Kd) for the streptavidin-biotin interaction lies in the range of 10–14 to 10–15 M, indicative of extraordinary affinity surpassing most antibody-antigen interactions. The tight binding is also thermodynamically driven by an exceptionally favorable free energy of binding (ΔG), with enthalpic and entropic contributions balanced to create a stable complex. (Hirsch et al., 2002)
Binding Kinetics and Temperature Stability
The association rate constants (kon) range between 3.0 × 106 and 4.5 × 107 M-1s-1, reflecting an exceptionally rapid binding event (Srisa-Art et al., 2008). These rapid kinetics are crucial for applications where transient complexes must be captured before they dissociate or diffuse away.
The melting temperature (Tm) hovers around 75°C (Hirsch et al., 2002), allowing for rigorous washing steps that remove nonspecific binders without disrupting streptavidin's tetrameric structure. This thermal resilience also supports processes that involve elevated temperatures, such as certain PCR-based assays or harsh chemical treatments. Stability studies often show that streptavidin maintains >90% of its biotin-binding capacity over extended storage at 4°C or after multiple freeze-thaw cycles.
Thermodynamic analyses indicate that the streptavidin-biotin interaction is not only strong but also exceptionally stable over time. Surface plasmon resonance (SPR) studies detect negligible dissociation events, underlining the quasi-irreversible nature of the binding (Gonzalez et al., 2000; Sano et al., 1997).
Natural Function and Distinctions from Avidin
In its native environment within Streptomyces avidinii, streptavidin acts as a biotin scavenger (Green, 1990). By sequestering this essential cofactor, the bacterium secures a metabolic advantage, ensuring adequate supply for carboxylation reactions. This natural function is mirrored in the laboratory to capture and immobilize biotinylated molecules with extraordinary selectivity. Avidin, found in egg white, is also a tetrameric biotin-binding protein. However, avidin is glycosylated and carries a higher isoelectric point, often resulting in more nonspecific interactions (Green, 1990; Hirsch et al., 2002). Streptavidin's non-glycosylated, more neutral character generally leads to cleaner backgrounds and more consistent performance across a variety of experimental conditions. Structural modifications and directed evolution studies on both proteins have shown that even subtle changes to surface residues can influence binding kinetics, specificity, and nonspecific adherence (Marttila et al., 1998).
Applications in Biotechnology and Analytical Workflows
Immunoassays, ELISA, and Signal Amplification
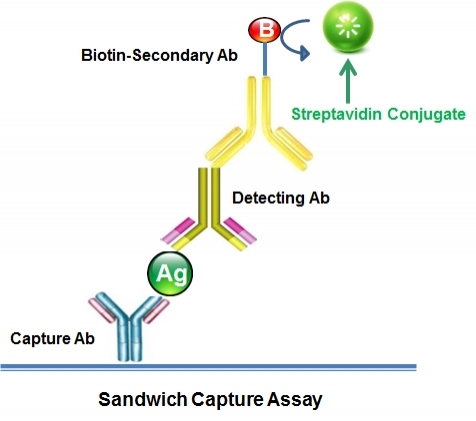
In ELISA and similar immunoassays, streptavidin-coated plates provide a stable, uniform platform for capturing biotinylated targets. Their structural resilience allows the use of stringent conditions—including high ionic strength buffers or guanidinium chloride—without damaging streptavidin's integrity, which in turn enables more thorough contaminant removal. This heightened tolerance to harsher protocols results in greater assay sensitivity, reproducibility, and fidelity.
Unlike enzymes or IgG-binding proteins, streptavidin is a pure binding protein with no intrinsic catalytic activity or immunoglobulin-binding domains. This neutral, nonspecific nature is advantageous: it can be easily coupled to secondary antibodies, creating streptavidin antibody conjugates that expand detection options. Streptavidin can also be linked to horseradish peroxidase (HRP), further enhancing signal amplification. Together, these attributes make streptavidin-based conjugates integral to versatile, high-sensitivity detection strategies in ELISA and related assays.
To create these streptavidin-coated ELISA plates, streptavidin is typically adsorbed onto polystyrene wells at a concentration of about 1–5 µg/mL in phosphate-buffered saline (pH ~7.4). Incubation for 1–2 hours at room temperature or overnight at 4°C allows streptavidin to bind to the plate surface. Unbound protein is then removed, and the plate is blocked—often with BSA or casein—to minimize nonspecific interactions. Following a final wash, the plates can be stored dry or in buffer, maintaining accessible biotin-binding sites that support subsequent assay steps.
Nucleic Acid Applications and Streptavidin Beads: Binding, Capture, and Elution
Streptavidin does not inherently bind non-biotinylated DNA. Instead, nucleic acids must be biotinylated to exploit streptavidin's extraordinary affinity. By incorporating biotinylated nucleotides, oligonucleotides, or adapters, researchers can selectively capture DNA or RNA fragments on streptavidin-coated surfaces. In next-generation sequencing (NGS) workflows, this approach improves library complexity, reduces off-target sequences, and facilitates targeted enrichment steps. Beyond NGS, single-molecule techniques such as single-molecule FRET, optical trapping, or atomic force microscopy leverage streptavidin's stable anchoring to investigate molecular interactions and conformational dynamics at unparalleled resolution (Gonzalez et al., 2000).
Streptavidin beads (magnetic or agarose) offer a practical, scalable platform for purifying biotinylated proteins, peptides, and nucleic acids. By simply adding a biotinylated target to a mixture containing streptavidin beads, researchers achieve rapid isolation with minimal contaminants. Specialized products, like Miltenyi's Streptavidin microbeads or various sized (0.6 µm, 1 µm and 3 µm) streptavidin-coated magnetic beads from AAT Bioquest, ensure consistent, high-activity binding surfaces. This versatility underpins complex protocols—such as immunoprecipitations, ribosome profiling, or chromatin purification—where specificity and purity are critical (Hofmann et al., 1980; Hirsch et al., 2002).
Once nucleic acids have been immobilized on streptavidin beads, controlled elution strategies enable their gentle release. Precise buffer conditions (e.g., altered pH or ionic strength) can weaken the streptavidin-biotin interaction enough to free intact nucleic acids. Even more effective is using desthiobiotin-labeled DNA instead of biotin. Desthiobiotin still binds streptavidin tightly but can be displaced by introducing free biotin under milder conditions (Hirsch et al., 2002). This approach preserves the structural and functional integrity of the DNA, ensuring its suitability for downstream applications like PCR amplification, sequencing, or cloning.
For non-nucleic acid targets or when desthiobiotin is not employed, additional methods exist to release biotin or its analogs from streptavidin. Harsh treatments, such as boiling in SDS, denature streptavidin and free bound biotin. However, these methods often destroy the protein's functionality. Gentler solutions include desthiobiotin (as mentioned above) and iminobiotin, which exhibits pH-dependent affinity: at alkaline pH, it binds tightly to streptavidin, while at slightly acidic pH, its affinity drops, enabling elution without protein denaturation (Hirsch et al., 2002; Hofmann et al., 1980). These reversible binding strategies represent alternatives within the broader streptavidin toolbox, allowing iterative selection cycles, multi-step purifications, and non-destructive sample recovery.
Engineered Tags: The Strep-tag and Twin-Strep-tag® Systems
To adapt streptavidin-based tools for protein purification under mild conditions, peptide affinity tags like the Strep-tag II were developed. Strep-tag II (WSHPQFEK) interacts with Strep-Tactin®, a genetically engineered streptavidin variant. Strep-Tactin® features altered amino acids in the biotin-binding site, offering roughly tenfold higher affinity for Strep-tag II compared to natural streptavidin. Under standard conditions, the Strep-tag II–Strep-Tactin® complex exhibits a dissociation constant (Kd) in the low micromolar range (~1 µM), allowing efficient purification and straightforward desthiobiotin-mediated elution (Schmidt et al., 2013; Skerra & Schmidt, 2000).
Building on this foundation, the Twin-Strep-tag® arranges two Strep-tag II motifs in tandem, dramatically enhancing affinity through avidity effects. The Twin-Strep-tag®–Strep-Tactin® interaction reaches the low nanomolar range, an improvement of two to three orders of magnitude over the single tag (Schmidt et al., 2013). This heightened affinity can result in higher purity, greater yield, and resistance to stringent wash steps, a boon in demanding purification schemes.
While Twin-Strep-tag® can still bind to streptavidin, the interaction is markedly weaker due to the absence of engineered mutations that optimize peptide binding. Thus, to fully exploit the ultrahigh affinity and its associated purification advantages, protocols typically employ Strep-Tactin® rather than native streptavidin. Nevertheless, rational protein engineering could tailor streptavidin variants to improve binding for Twin-Strep-tag®, illustrating streptavidin's modifiability and the growing complexity of streptavidin-based systems.
Evolving Toolkits and Integrative Approaches
While the biotin–streptavidin system remains foundational, its role has become more nuanced as research demands intensify. Today's molecular toolkit integrates orthogonal approaches that complement or extend beyond the classic biotin–streptavidin interaction, enabling enhanced specificity, environmental responsiveness, multiplexed detection, and finer control over capture and release steps.
Nucleic acid aptamers, derived through iterative selection processes like SELEX (Ellington and Szostak, 1990), can match or surpass antibody-like affinities, yet remain stable under a broad range of conditions. They may function independently or in parallel with biotin–streptavidin, thus facilitating assays where multiple binding events operate side-by-side without interference. Similarly, engineered protein scaffolds (Gilbreth and Koide, 2012) employ stable protein folds as customizable frameworks for designing high-affinity, selective binding surfaces tolerant of harsh or nontraditional assay conditions. Likewise, synthetic receptor–ligand pairs (De Los Santos et al., 2020; Liu et al., 2017) introduce tunable binding that responds to subtle shifts in pH or ionic strength, enabling reversible capture, gentle elution, and less disruptive recovery of target molecules compared to the sometimes harsh treatments needed to detach biotin from streptavidin.
Amid these expanding options, the biotin–streptavidin system itself continues to evolve, incorporating refinements that preserve its core advantages while increasing adaptability. Improving conjugation strategies to achieve precise fluorophore density mitigates self-quenching and background fluorescence, producing sharper signals even in demanding assays. Coupling streptavidin with advanced fluorophores (e.g., iFluor®, mFluor™) yields brighter, more photostable complexes suitable for intricate, multi-wavelength detection formats. To address challenges in distinguishing biotin from proteins or nucleic acids, reagents like ReadiView™ introduce hydrophilic spacers and quantifiable color tags, ensuring uniform labeling and consistent, reproducible outcomes. These incremental improvements amplify the biotin–streptavidin framework's inherent strengths.
As technology continues to progress, streptavidin's integration into even more sophisticated workflows becomes routine. Microfluidic devices rely on streptavidin-coated channels to immobilize biotinylated probes for rapid, on-chip assays that conserve reagents and enable real-time analysis. Single-molecule localization microscopy (SMLM) and super-resolution techniques exploit the stable attachment of biotinylated markers through streptavidin to achieve spatial resolutions on the nanometer scale. In proteomics, the combination of biotin tagging with streptavidin-based enrichment selectively isolates subsets of proteins for in-depth mass spectrometric characterization. Additionally, in vivo biotinylation approaches (Barat and Wu, 2007; Emerman et al., 2010) allow for metabolic incorporation of biotin into newly expressed proteins, simplifying downstream purification and detection—even from complex biological contexts.
Further Reading
View All
Barat, B., and Wu, A. M. "Metabolic Biotinylation of Recombinant Antibody by Biotin Ligase Retained in the Endoplasmic Reticulum." Biomolecular Engineering, vol. 24, 2007, pp. 283–91.
Chaiet, L., and Wolf, G. "The Properties of Streptavidin, a Biotin-Binding Protein Produced by Streptomycetes." Archives of Biochemistry and Biophysics, vol. 106, no. 1, 1964, pp. 1–5.
De Los Santos, C., et al. "Engineering Orthogonal Ligand–Receptor Pairs from the Streptavidin–Biotin System." Protein Engineering, Design & Selection, vol. 33, no. 2, 2020, pp. 41–49.
Ellington, A. D., and Szostak, J. W. "In Vitro Selection of RNA Molecules That Bind Specific Ligands." Nature, vol. 346, 1990, pp. 818–822.
Emerman, A. B., et al. "Compartment-Restricted Biotinylation Reveals Novel Features of Prion Protein Metabolism in Vivo." Molecular Biology of the Cell, vol. 21, 2010, pp. 4325–37.
Gilbreth, R. N., and Koide, S. "Structural Insights for Engineering Binding Proteins Based on Non-Antibody Scaffolds." Current Opinion in Structural Biology, vol. 22, no. 4, 2012, pp. 413–20.
Gonzalez, M., et al. "Structure–Function Analysis of the Streptavidin–Biotin Interaction." Biophysical Journal, vol. 79, no. 6, 2000, pp. 3131–3140.
Green, N. M. "Avidin and Streptavidin." Methods in Enzymology, vol. 184, Academic Press, 1990, pp. 51–67.
Gosling, James P. ELISA: Theory and Practice. Academic Press, 1990.
Hermanson, G. T. Bioconjugate Techniques. 2nd ed., Academic Press, 2008.
Hirsch, J., et al. "Easily Reversible Desthiobiotin Binding to Streptavidin, Avidin, and Other Biotin-Binding Proteins: Uses for Protein Labeling, Detection, and Isolation." Analytical Biochemistry, vol. 308, 2002, pp. 343–357.
Hofmann, K., et al. "Iminobiotin Affinity Columns and Their Application to Retrieval of Streptavidin." Proceedings of the National Academy of Sciences USA, vol. 77, 1980, pp. 4666–8.
Laitinen, O. H., et al. "Structural Origins of High-Affinity Biotin Binding in Streptavidin and Avidin." Proceedings of the National Academy of Sciences, vol. 103, no. 17, 2006, pp. 6295–6300.
Liu, W., et al. "Synthetic Mimics of Biotin/(Strept)avidin." Bioconjugate Chemistry, 2017, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5423825/.
Luong, J. H. T., and Vashist, S. K. "Chemistry of Biotin–Streptavidin and the Growing Concern of an Emerging Biotin Interference in Clinical Immunoassays." ACS Omega, vol. 5, no. 1, 2020, pp. 10–18.
Marttila, A. T., et al. "Engineering of Chicken Avidin: A Progressive Series of Reduced Charge Mutants." FEBS Letters, vol. 441, no. 2, 1998, pp. 313–317.
Sano, T., Vajda, S., Smith, C. L., and Cantor, C. R. "Engineering of Streptavidin Variants with Altered Ligand-Binding Kinetics." Biochemical and Biophysical Research Communications, vol. 233, no. 3, 1997, pp. 764–767.
Schmidt, T. G. M., et al. "Development of the Twin-Strep-Tag® and Its Application for Purification of Recombinant Proteins from Cell Culture Supernatants." Protein Expression and Purification, vol. 92, no. 1, 2013, pp. 54–61.
Skerra, A., & Schmidt, T. G. M. "Use of the Strep-tag and Streptavidin for Detection and Purification of Recombinant Proteins: Molecular Origins and Applications." Biomolecular Engineering, vol. 16, no. 1–4, 2000, pp. 79–86.
Srisa-Art, M., et al. "Monitoring of Real-Time Streptavidin–Biotin Binding Kinetics Using Droplet Microfluidics." Analytical Chemistry, vol. 80, no. 18, 2008, pp. 7063–7067.
Wilchek, M., and Bayer, E. A. "Applications of Avidin–Biotin Technology: Literature Survey." Methods in Enzymology, vol. 184, Academic Press, 1990, pp. 14–45.
Citations
View all 18 citations: Citation Explorer
Expanding the Repertoire of Poly (Oxazoline) Functionality
Authors:
Garcia, Joseph Angel
SERS-based cascade amplification bioassay protocol of miRNA-21 by using sandwich structure with biotin-streptavidin system
Authors:
Liang, Z., Zhou, J., Petti, L., Shao, L., Jiang, T., Qing, Y., Xie, S., Wu, G., Mormile, P.
Journal:
Analyst (2019): 1741-1750
Size effects of magnetic beads in circulating tumour cells magnetic capture based on streptavidin-biotin complexation
Authors:
Li, F., Xu, H., Sun, P., Hu, Z., Aguilar, Z. P.
Journal:
IET Nanobiotechnol (2019): 6-11
Streptavidin interfacing as a general strategy to localize fluorescent membrane tension probes in cells
Authors:
Goujon, A., Strakova, K., Sakai, N., Matile, S.
Journal:
Chem Sci (2019): 310-319
Erroneous thyroid and steroid hormones profile due to anti-streptavidin antibodies
Authors:
Bayart, J. L., Favresse, J., Melnik, E., Lardinois, B., Fillee, C., Maiter, D., Gruson, P. D.
Journal:
Clin Chem Lab Med (2019): se name="16885.enl" path="C:\Website\Referenc
References
View all 195 references: Citation Explorer
Overexpression of CXCR2 predicts poor prognosis in patients with colorectal cancer.
Authors:
Zhao, Jingkun and Ou, Baochi and Feng, Hao and Wang, Puxiongzhi and Yin, Shuai and Zhu, Congcong and Wang, Shenjie and Chen, Chun and Zheng, Minhua and Zong, Yaping and others, undefined
Journal:
Oncotarget (2017)
Cadherin-12 enhances proliferation in colorectal cancer cells and increases progression by promoting EMT
Authors:
Ma, Junjun and Zhao, Jingkun and Lu, Jun and Wang, Puxiongzhi and Feng, Hao and Zong, Yaping and Ou, Baochi and Zheng, Minhua and Lu, Aiguo
Journal:
Tumor Biology (2016): 1--12
Transplantation of RADA16-BDNF peptide scaffold with human umbilical cord mesenchymal stem cells forced with CXCR4 and activated astrocytes for repair of traumatic brain injury
Authors:
Shi, W and Huang, CJ and Xu, XD and Jin, GH and Huang, RQ and Huang, JF and Chen, YN and Ju, SQ and Wang, Y and Shi, YW and others, undefined
Journal:
Acta Biomaterialia (2016): 247--261
The migration and differentiation of hUC-MSCsCXCR4/GFP encapsulated in BDNF/chitosan scaffolds for brain tissue engineering
Authors:
Huang, Chuanjun and Zhao, Longxiang and Gu, Jun and Nie, Dekang and Chen, Yinan and Zuo, Hao and Huan, Wei and Shi, Jinlong and Chen, Jian and Shi, Wei
Journal:
Biomedical Materials (2016): 035004
Antiprothrombin antibodies in a patient with secondary antiphospholipid syndrome and bleeding
Authors:
Gonzalez Leon R, Garcia Hern and ez FJ, Castillo Palma MJ, Sanchez Roman J.
Journal:
Med Clin (Barc) (2011): 668
Physical properties
| Solvent | Water |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12171501 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 11, 2026
