Screen Quest™ Fluorimetric Fatty Acid Uptake Assay Kit
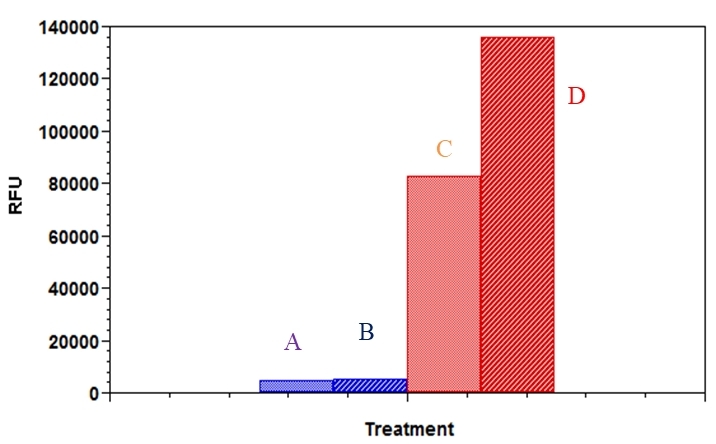
Fatty acid uptake is an important therapeutic target for the treatment of many human diseases such as obesity, type 2 diabete and hepatic steatosis. The Screen Quest™ Fluorimetric Fatty Acid Uptake Assay Kit provides a simple and sensitive method for the measurement of fatty acid uptake in cells containing fatty acid transporters. The kit uses a proprietary dodecanoic acid fluorescent fatty acid substrate. This fatty acid uptake assay kit can be performed on any fluorescence microplate reader with a bottom-read mode or FITC channel. The assay can be performed in 96-well or 384-well microtiter plates in a simple mix-and-read procedure, and easily adapted for high throughput screening applications.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 36385 | 100 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 485 nm |
| Emission | 515 nm |
| Cutoff | 495 nm |
| Recommended plate | Black wall/clear bottom |
| Instrument specification(s) | Bottom read mode |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 30, 2026
