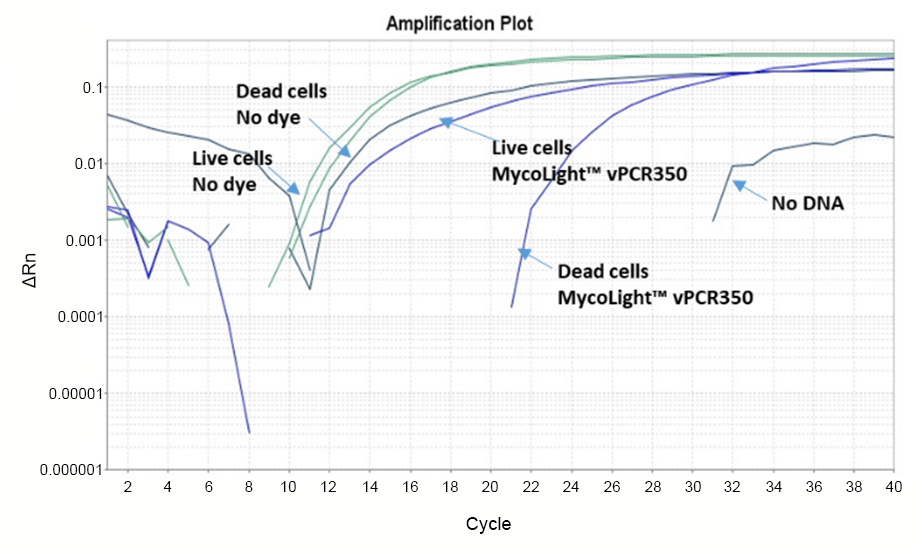
MycoLight™ vPCR350 is a novel non-fluorescent DNA modifier specifically designed for viability PCR (vPCR) targeting microorganisms such as bacteria, eukaryotes, viruses, and fungi. It represents a significant advancement over the widely used viability dye PMA (propidium monoazide), offering enhanced performance and specificity. Like PMA, MycoLight™ vPCR350 is a photo-reactive dye that strongly binds to DNA. Upon exposure to visible light, the dye covalently attaches to the DNA, effectively inhibiting its amplification by PCR. A notable feature of MycoLight™ vPCR350 is its designed impermeability to cell membranes. This unique property ensures that only dead cells with compromised membranes, are susceptible to DNA modification by the dye thereby eliminating PCR amplification of dead cell DNA and ensuring precise discrimination between live and dead bacteria. While PMA is effective at distinguishing between live and dead bacteria, it does not entirely prevent PCR products from dead cell DNA, potentially leading to false positive results. MycoLight™ vPCR350 is compatible with all existing PMA protocols. Its superior performance and reliability make it an ideal choice for microbial identification by real-time PCR (qPCR), offering a highly specific and efficient means of detection without the need to cultivate the target organisms.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 24210 | 1 mg | Price |
| Solvent | DMSO |
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12171501 |
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
