Cell Meter™ Glucose Uptake Imaging Kit
Red Fluorescence
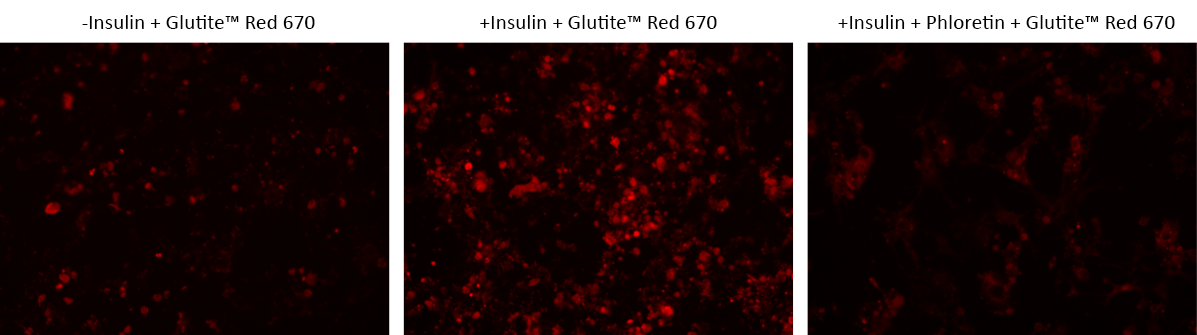
Glucose metabolism is a primary source of cellular energy and biomaterials for maintaining cell homeostasis. Surplus glucose is stored as glycogen in muscular and hepatic tissues, with subsequent release into the bloodstream as needed. The GLUT family of transporter proteins facilitates the process of glucose uptake under the regulation of multiple mechanisms, including the modulation by hormones and growth factors like insulin. Notably, cancer cells exhibit a marked propensity for increased glucose uptake and metabolic activity, engaging in aerobic glycolysis to sustain their accelerated proliferation rates. Compounds that hinder glucose uptake in cancer cells exhibit discernible anti-cancer effects. The Cell Meter™ Glucose Uptake Imaging Kit provides a simple and direct method for quantifying glucose uptake across diverse cellular contexts. This kit employs Glutite™ Red 670, a cell-permeable fluorescent glucose tracer, enabling precise molecular sensing and bioimaging via GLUT transporters. The robust red fluorescence emitted by Glutite™ Red 670 (Ex/Em = 651/670 nm) correlates proportionally with cellular glucose uptake, offering a valuable means for quantification via both fluorescence microscopy and flow cytometry techniques.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 23501 | 100 Tests | Price |
Physical properties
| Solvent | DMSO |
Spectral properties
| Extinction coefficient (cm -1 M -1) | 250000 |
| Excitation (nm) | 651 |
| Emission (nm) | 670 |
| Quantum yield | 0.27 1 , 0.42 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microscope | |
| Excitation | Cy5 Filter Set |
| Emission | Cy5 Filter Set |
| Recommended plate | Black wall/clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 15, 2026

