Cell Meter™ Fluorimetric Intracellular Total ROS Activity Assay Kit
Green Fluorescence
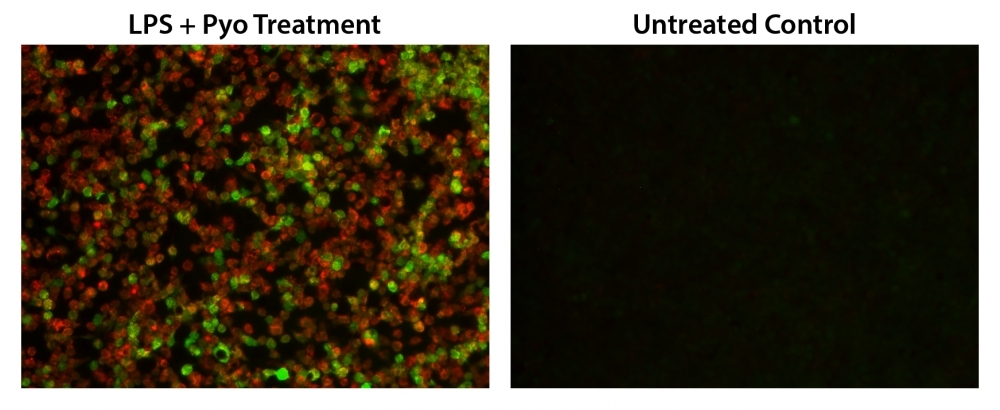
Reactive oxygen species (ROS) are natural byproducts of the normal metabolism of oxygen and play important roles in cell signaling. However, during oxidative stress-related states, ROS levels can increase dramatically. The accumulation of ROS results in significant damage to cell structures. The role of oxidative stress in cardiovascular disease, diabetes, osteoporosis, stroke, inflammatory diseases, a number of neurodegenerative diseases and cancer has been well established. The ROS measurement will help to determine how oxidative stress modulates varied intracellular pathways. Cell Meter™ Fluorimetric ROS Assay Kit uses our unique ROS sensor to quantify ROS in live cells. ROS Green is cell-permeable. It generates the green fluorescence when it reacts with ROS. The kit is an optimized "mix and read" assay format that is compatible with HTS liquid handling instruments. The Cell Meter™ Fluorimetric ROS Assay Kit provides a sensitive, one-step fluorimetric assay to detect intracellular ROS in live cells with one hour incubation. The assay can be performed in a convenient 96-well or 384-well microtiter-plate format and easily adapted to automation without a separation step. Its signal can be easily read using either a fluorescence microplate reader or a fluorescence microscope.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22900 | 200 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microscope | |
| Excitation | FITC filter |
| Emission | FITC filter |
| Recommended plate | Black wall/clear bottom |
| Fluorescence microplate reader | |
| Excitation | 490 nm |
| Emission | 525 nm |
| Cutoff | 515 nm |
| Recommended plate | Black wall/clear bottom |
| Instrument specification(s) | Bottom read mode |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 20, 2026
