Cell Meter™ Fluorimetric Intracellular Peroxynitrite Assay Kit
Green Fluorescence
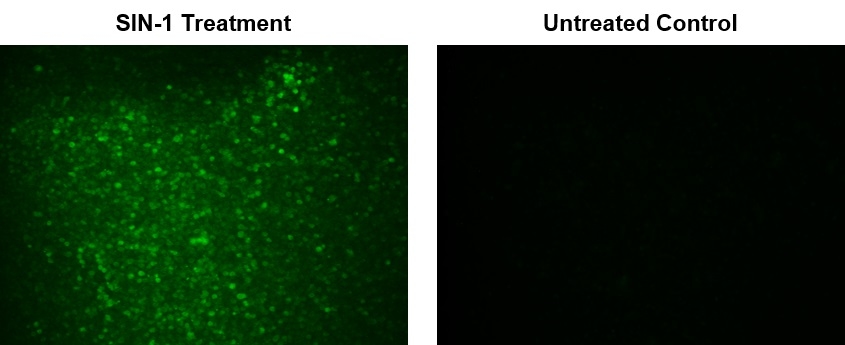
Peroxynitrite (ONOO-) is a strong oxidizing species and a highly active nitrating agent. Peroxynitrite is formed from the reaction between superoxide radicals and nitric oxide generated in cells. It can cause damages to a wide array of biomolecules including proteins, enzymes, lipids and nucleic acids, eventually contributing to cell death. Meanwhile, peroxynitrite can also have protective activities in vivo by contributing to host-defense responses against invading pathogens. Therefore, peroxynitrite is an essential biological oxidant involved in a board range of physiological and pathological processes. Due to its extremely short half-life and low steady-state concentration, it has been challenging to detect and understand the role of peroxynitrite in biological systems. AAT Bioquest's DAX-J2™ PON Green has been developed to address this unmet need. It provides a sensitive tool to monitor ONOO- level in living cells. AAT Bioquest's DAX-J2™ PON Green specifically reacts with intercellular ONOO- to generate a bright green fluorescent product. It can be used in fluorescence imaging, flow cytometry and fluorescence microplate reader-based assays.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 16315 | 100 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microscope | |
| Excitation | 490 nm |
| Emission | 530 nm |
| Recommended plate | Black wall/clear bottom |
| Instrument specification(s) | FITC Filter Set |
| Fluorescence microplate reader | |
| Excitation | 490 nm |
| Emission | 530 nm |
| Cutoff | 515 nm |
| Recommended plate | Black wall/clear bottom |
| Instrument specification(s) | Bottom read mode |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 29, 2026
