Protein Tags

Staining of microtumors with DIR-RGD-NP. Fluorescence spectral images from dissected intestines and the attached mesentery. Images shown are from mCherry channel (red; left column) and DIR channel (green; middle column). The merged images (right column) demonstrate the best colocalization of mCherry and DIR signals (white arrow) in animals that received DIR-RGD-NP. Source: Novel approach for the detection of intraperitoneal micrometastasis using an ovarian cancer mouse model by Alvero et al., Scientific Reports, Jan. 2017.
Viral proteins are great candidates for protein tags as they are unique. The distinctive nature of viral proteins helps minimize and mitigate any potential issues with cross-reactivity or antibodies, as well as the copurification of nontarget proteins. Though tagging can be performed by vector cloning as mentioned, CRISPR/Cas9 gene editing has also become a widely used method for use in endogenous proteins. It is important to note that the addition of a protein tag may result in partial and/or complete loss of function, which may warrant the need for a complementation study or side-by-side analysis of non-target proteins.
Table of Contents
- Common Varieties of Protein Tags
- 1.1 Polyhistidine (His-tag or His6)
- 1.2 Glutathione-S-transferase (GST) Tags
- 1.3 Tandem Affinity Purification (TAP) Tags
- 1.4 Maltose-Binding Protein (MBP) Tags
- 1.5 Intein-Chitin Binding Domain (intein-CBD) Tag
- 1.6 Calmodulin Binding Protein (CBP) Tags
- 1.7 HiBiT Peptide Tag
- 1.8 Epitope Tags
- 1.9 Fluorescent Protein Tags
- Product Ordering Information
- References
Common Varieties of Protein Tags
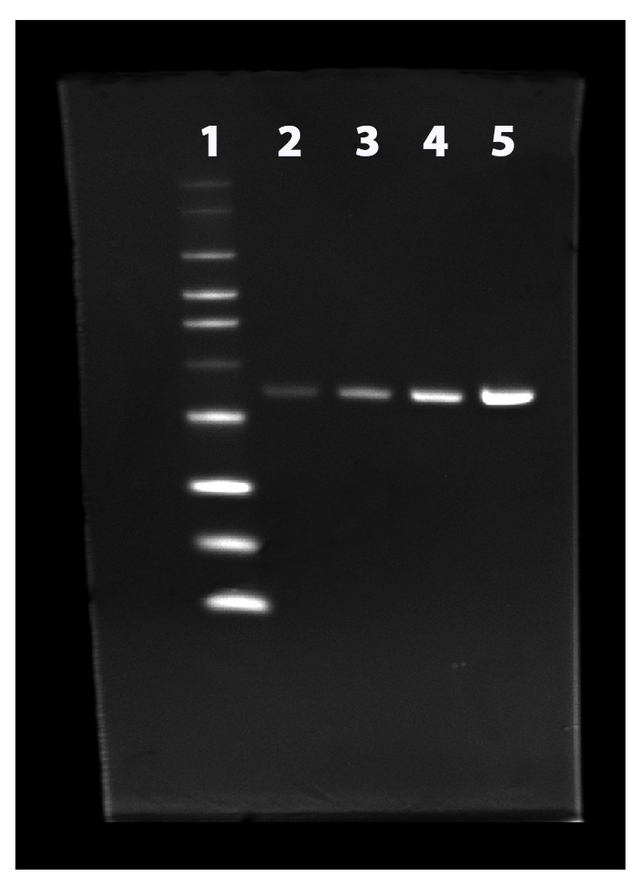
Two-fold dilution series of His-tagged annexin V were separated on a NuPAGE® 4-12% Bis-Tris gel and stained with the ProLite™ His-Tag Protein Gel Staining Kit according to standard protocols. Lane 1: His-tagged protein ladder, Lane 2 to 5: two-fold dilution of His-tagged annexin V.
Polyhistidine (His-tag or His6)
His-tags are composed of 6-8 consecutive histidine residues, though may vary in length between 2-10 residues. Unlike epitope tags, which when duplicated can increase the tag size quickly, modifying the length of a His tract does not greatly alter the tag size. The relatively small size of His-tags not only makes integration into expression vectors extremely easy, but also less likely than other tags to affect function. His-tagged proteins are usually purified using immobilized nickel (Ni2+), cobalt (Co2+) or zinc (Zn2+) ions through metal affinity chromatography . Copper (Cu2+), Calcium (Ca2+), and Iron (Fe3+) have also been experimentally used, though empirical determination of the most effective transition metal ion should be a consideration. His-tags do not form secondary structures to bind to their substrate, so purification can even be performed even under denaturing conditions, which is exceedingly useful if protein solubility may be an issue.
A secondary elution step may be added, if desired, through the use of EDTA or imidazole. Imidazole elution has been reported often as it is relatively mild and can help preserve the immunogenicity of His-fusion proteins. Purification of a highly-expressed His-fusion proteins can lead to relatively pure protein (>80%) in one chromatographic step, though purification from insect and mammalian cells may lead to significant background binding to immobilized metal ions due to the higher percentage of His residues that these cells naturally contain. Due to this factor, anti-polyhistidine antibodies are widely used for mammalian cell samples. Several commercial kits are available for purifying His-tagged proteins in native, denaturing, or hybrid conditions.
Table 1. HIS Lite™ NTA-Ni conjugates
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 12610 | HIS Lite™ Cy3 Bis NTA-Ni Complex | 1 mg |
| 12613 | HIS Lite™ Cy5 Bis NTA-Ni Complex | 1 mg |
| 12615 | HIS Lite™ OG488-Tris NTA-Ni Complex | 100 µg |
| 12617 | HIS Lite™ iFluor® 568 Tris NTA-Ni Complex | 100 µg |
| 12618 | HIS Lite™ iFluor® 647 Tris NTA-Ni Complex | 100 µg |
| 12619 | HIS Lite™ Cy5 Tris NTA-Ni Complex | 100 µg |
| 12620 | HIS Lite™ Cy3 Tris NTA-Ni Complex | 100 µg |
| 12621 | HIS Lite™ Cy3B Tris NTA-Ni Complex | 100 µg |
Table 2. HIS tag antibodies
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| V102000 | Purified Mouse Anti-6xHIS Tag Antibody *Monoclonal* | 0.1 mg |
| V102005 | Purified Rabbit Anti-6xHIS Tag Antibody *Polyclonal* | 0.1 mg |
| V102010 | Biotin Rabbit Anti-6xHIS Tag Antibody *Polyclonal* | 0.1 mg |
| V102015 | FITC Rabbit Anti-6xHIS Tag Antibody *Polyclonal* | 0.1 mg |
| V102020 | HRP Rabbit Anti-6xHIS Tag Antibody *Polyclonal* | 0.1 mg |
Glutathione-S-transferase (GST) Tags

Immunohistochemistry analysis of paraffin-embedded human lung carcinoma tissue using a microsomal Glutathione S-Transferase 3 (MGST3) antibody.
When using GST tags, first proteins are purified using beads coated glutathione (Glu), its substrate, to which it binds with high affinity. Next, recombinant proteins are eluted with a buffer containing free Glu. As the protein attaches to the GST tag, its native state will be altered. It is vital that the GST tag be correctly folded for successful binding to Glu, though even if the GST tag refolds, the protein of interest may not.
If the expressed protein has aggregated into an inclusion body, affinity purification using Glu becomes problematic. An additional step may be required to cleave the target protein from the GST tag, to reduce possible functional interference. GST-tagged proteins can be detected by a colorimetric assay with the GST substrate of 1-chloro-2,4-dinitrobenzene (CDNB) or with anti-GST antibodies.
| Inhibitor Datasets: | ||
Tandem Affinity Purification (TAP) Tags

Protein A agarose beads have physical and chemical properties that enable them to be used in a variety of affinity purification procedures. Protein A contains four high-affinity binding sites interacting with the Fc region of IgG-class antibodies from selected mammalian species.
The major advantage to this method is that, due to the two purification steps involved, nonspecific background binding is reduced. Though 20-30% of a protein of interest can be obtained using this method, TAP tags do not work well in higher eukaryotes. The large tag size of the tag, approximately 21 kDa, can disturb protein function and CBP may interfere with calcium signaling. These issues have led to the creation of over 30 TAP tag variants.
Maltose-Binding Protein (MBP) Tags
MBP tags bind to amylose agarose and are commonly used to increase the expression level and solubility of fusion proteins. The relatively large size of MBP tags, roughly 40-45 KDa, may affect protein function and like GST fusion proteins, high-level expression of MBP fusion proteins may result in an over-accumulation of insoluble protein aggregates in inclusion bodies. MBP fusion proteins can be purified by affinity chromatography on a cross-linked amylose resin, and then must be eluted using a buffer containing free maltose before cleavage. Amylose resins are commercially available, but are affected by amylase activity in crude cell lysates and can be reused only 3-5 times. Amylose affinity chromatography is also not well suited for use with denaturing or reducing agents. Multiple anti-MBP antibodies are available to aid in the detection of MBP fusion proteins.
Intein-Chitin Binding Domain (intein-CBD) Tag
The intein-CBD tag is a combination of a protein self-splicing element (intein) with a chitin-binding domain that allows for the purification of a native recombinant protein without the need for a protease. CBD fusion proteins are purified by affinity chromatography on chitin resin, which is commercially available and can be regenerated up to 5 times. Chitin affinity chromatography, like MBP tags, is not amenable to denaturing reagents such as urea or guanidine hydrochloride.
Calmodulin Binding Protein (CBP) Tags

Immunohistochemistry analysis of paraffin-embedded human brain tissue using Calmodulin (Ab-79/81) antibody.
Table 3. Anti-Calmodulin Binding Peptide Antibodies
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| V102035 | Purified Rabbit Anti-Calmodulin Binding Peptide Antibody *Polyclonal* | 0.1 mg |
| V102040 | Biotin Rabbit Anti-Calmodulin Binding Peptide Antibody *Polyclonal* | 0.1 mg |
| V102045 | FITC Rabbit Anti-Calmodulin Binding Peptide Antibody *Polyclonal* | 0.1 mg |
| V102050 | HRP Rabbit Anti-Calmodulin Binding Peptide Antibody *Polyclonal* | 0.1 mg |
HiBiT Peptide Tag
HiBiT tags, about 11 amino acids in length, may also be used to and offer a very small tag length. On its own, HiBiT is not bioluminescent, though upon the addition of a detection reagent containing the complementary LgBiT protein is added, luminescence is produced. As bioluminescent assays are increasingly sensitive and facilitate detection down to nanomolar levels, the luminescent signal is quantifiable and can be measured using a standard luminometer.
Note: If LgBiT is expressed intracellularly, it is possible to kinetically detect and quantify HiBiT-tagged protein abundance in live cells.
Epitope Tags
Epitope tags are short peptide sequences, generally smaller than affinity tags, which are readily recognized by antibodies. The inherent small size of epitope tags mean they are normally unlikely to have a serious effect on the structure of the targeted fusion protein. Epitope tags are widely used for the detection of fusion proteins in vitro and in cell culture. Several epitope tags may be added to a protein in combination to generate larger tags that can be detected by antibodies to offer higher sensitivity.
Epitope tags can be purified using immobilized primary antibodies and specificity can be increased through the employment of an enzyme-linked secondary antibody. A variety of monoclonal antibodies (mAbs) are commercially available for these tags, making them particularly suitable for Western blotting, immunofluorescence, or immunoprecipitation experiments. Antibody affinity chromatography often involves a pH elution which frequently affects the properties of the fusion protein. Antibody affinity, as well, will employ the use of a resin that limits the possibility of reuse. For these reasons, epitope tags are typically not the first choice when the main goals are high-level expression and fusion protein purification.
Many commercially available epitope tags exist, one being the MYC Tag. The MYC tag is derived from the human c-myc proto-oncogene product, recognized by numerous commercial antibodies, and can be added to a protein using recombinant DNA technology. MYC Tags have been reported for use with affinity chromatography and for isolating protein complexes with more than 1 subunit.
Table 4. c-Myc tag Antibodies
The first epitope tag to be characterized, the FLAG tag, may also be used in recombinant DNA technology, for affinity chromatography, and for isolating protein complexes with multiple subunits. The FLAG tag is more hydrophilic than other similar epitope tags, and does not denature or inactivate the fusion protein. A third common epitope tag includes the human influenza hemagglutinin (HA) tag. This tag is derived from the surface glycoprotein that facilitates the ability of the influenza virus to infect its host, and is used mostly as a general epitope tag in expression vectors.
Fluorescent Protein Tags
Fluorescent tags have become the go-to protein tagging method to follow the abundance, localization, and/or movement of a protein of interest using microscopy or flow cytometry. Fluorescent tags are non-toxic, which makes them useful for detecting proteins in live as well as in fixed cells. Many times, fluorescent tags are used in combination with a secondary infrared extrinsically fluorescent protein. When choosing the appropriate fluorescent tag, there are a number of relevant parameters. It is important to consider whether a fluorescent tag may affect the behavior of the fused protein. Normally, it's difficult to predict (sometimes even impossible) if a fluorescent tag will affect protein function. Some fluorescent tags suffer from aggregation because, intrinsically, many fluorescent tags have been engineered from dimeric or tetrameric proteins, and often retain some residual aggregation tendency. There do exist, however, commercially available assays to address this specific problem.
The spectral properties, or color, of the fluorescent tag is determined by its excitation and emission spectra. If more than 1 fluorescent protein is desired in an experiment, emitted color output must be of high consideration to allow for easy detection. The most popular fluorescent protein tags are GFP and RFPs, although a wide variety of options exist. The brightness of the fluorescent tag must also be a consideration. The brightness is measured by the product of the extinction coefficient (how effectively the fluorophore absorbs light) and the quantum yield (what fraction of absorbed photons produce fluorescence). Photostability of your fluorescent tag must also be taken into account because, when excited, fluorescent molecules may undergo side reactions that lead to the destruction of the fluorophore. This destruction could result in the loss of fluorescent yield over time, a phenomena known as photobleaching. How quickly photobleaching occurs is often variable, and depends nonlinearly on the excitation light and the length of illumination.
| Datasets: | ||
Product Ordering Information
Table 5. Polyhistidine Tag (His-Tag) Kits
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 18010 | ProLite™ His-Tag Protein Gel Staining Kit *Green Fluorescence* | 10 Gels |
Table 6. Building blocks for developing polyHIS probes
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 12633 | Tris NTA succinimidyl ester | 1 mg |
| 12634 | Tris NTA maleimide | 1 mg |
| 12635 | Tris NTA azide | 1 mg |
| 12636 | Tris NTA alkyne | 1 mg |
| 12637 | Tris NTA amine | 1 mg |
Table 7. Ordering Info for Amplite Glutathione Kits (P.O.T) Products
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 10055 | Amplite® Fluorimetric Glutathione Assay Kit *Green Fluorescence* | 200 Tests |
| 10056 | Amplite® Fluorimetric Glutathione GSH/GSSG Ratio Assay Kit *Green Fluorescence* | 200 Tests |
| 10060 | Amplite® Rapid Fluorimetric Glutathione GSH/GSSG Ratio Assay Kit *Green Fluorescence* | 200 Tests |
Table 8. Protein A for Affinity Purification
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 11041 | MegaWox™ polyHRP-Protein A Conjugate | 1 mg |
| 55000 | Protein A-Agarose Resin | 1 mL |
| 55005 | Protein A-Agarose Resin | 5 mL |
