mFluor™ UV375 SE
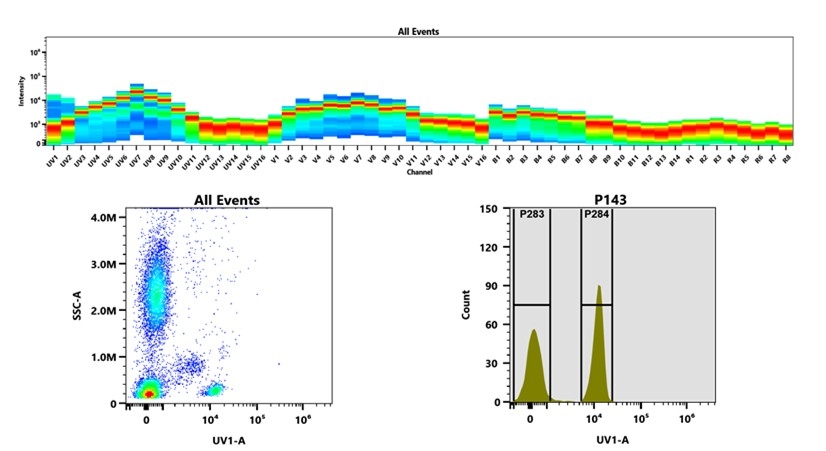
AAT Bioquest's mFluor™ dyes are developed for multicolor flow cytometry-focused applications. These dyes have large Stokes Shifts, and can be well excited by the laser lines of flow cytometers (e.g., 355 nm, 405 nm, 488 nm and 633 nm). mFluor™ UV375 (MFUV375) dyes have fluorescence excitation and emission maxima of ~355 nm and ~375 nm respectively. These spectral characteristics make them an excellent alternative to BD Biosciences' BUV395. mFluor™ UV375 dyes are well excited by UV laser of 355 nm with emission centered around 375 nm, which perfectly matches the filter set of 379/28 nm used for BUV 395. In contrast to polymer-based BUV 395 dye, MFUV375 is a small organic molecule that can be readily conjugated to antibodies. It is significantly dimmer than BUV395, might be used for brighter markers (such as CD4).

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 1135 | 1 mg | Price |
Physical properties
| Molecular weight | 782.95 |
| Solvent | DMSO |
Spectral properties
| Absorbance (nm) | 342 |
| Correction factor (260 nm) | 0.116 |
| Correction factor (280 nm) | 0.139 |
| Extinction coefficient (cm -1 M -1) | 40000 1 |
| Excitation (nm) | 351 |
| Emission (nm) | 387 |
| Quantum yield | 0.94 1 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12171501 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 21, 2026

